- EN - English
- CN - 中文
Preparation of Chromatin Fragments From Human Cells for Cryo-EM Analysis
用于冷冻电镜分析的人源染色质片段制备
发布: 2025年10月20日第15卷第20期 DOI: 10.21769/BioProtoc.5472 浏览次数: 1947
评审: Munenori IshibashiAnonymous reviewer(s)

相关实验方案

相关常规和超分辨率光激活定位显微镜 (PALM) 成像来表征活哺乳动物细胞的染色质结构和动力学
Dushyant Mehra and Elias M. Puchner
2023年10月20日 2511 阅读
一种从蚊子脂肪体中高质量分离单细胞核用于RNA测序的优化方法
Stephanie Serafim de Carvalho [...] Carolina Barillas-Mury
2026年01月05日 199 阅读
Abstract
Eukaryotic genomic DNA is packaged into chromatin, which plays a critical role in regulating gene expression by dynamically modulating its higher-order structure. While in vitro reconstitution approaches have offered valuable insights into chromatin organization, they often fail to fully capture the native structural context found within cells. To overcome this limitation, we present a protocol for isolating native chromatin fragments from human cells for cryo-electron microscopy (cryo-EM) analysis. In this method, chromatin from formaldehyde-crosslinked human HeLa S3 nuclei is digested with micrococcal nuclease (MNase) to generate mono- and poly-nucleosome fragments. These fragments are subsequently fractionated by sucrose-gradient ultracentrifugation and prepared for cryo-EM. The resulting chromatin fragments retain native-like nucleosome–nucleosome interactions, facilitating structural analyses of chromatin organization under near-physiological conditions.
Key features
• Chemical crosslinking preserves the native nucleosome–nucleosome interactions in chromatin fragments.
• Optimal MNase digestion conditions efficiently solubilize chromatin into mono- and poly-nucleosome fragments for cryo-EM analysis.
• This protocol may be adaptable to other types of cells.
Keywords: Native chromatin (天然染色质)Graphical overview
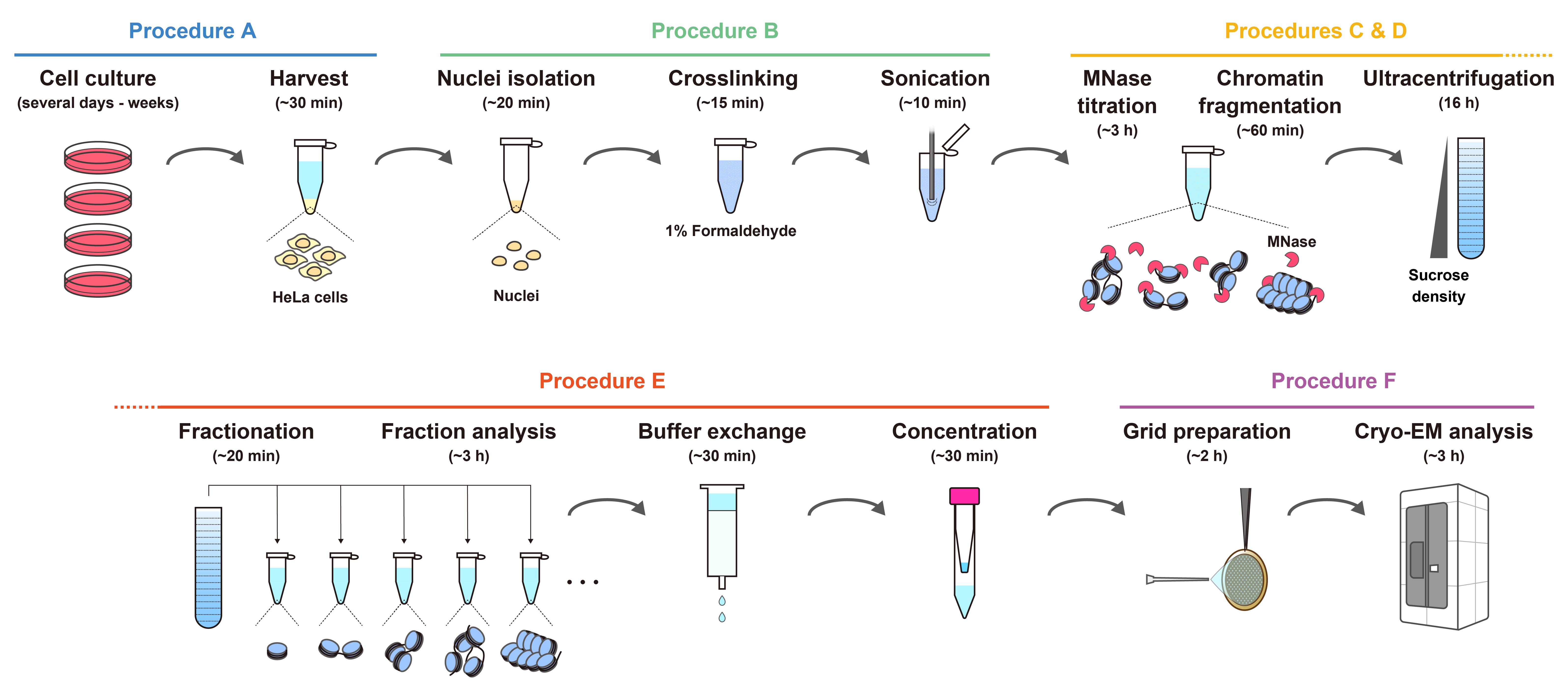
Workflow for the preparation of chromatin fragments from HeLa cells for cryo-EM analysis. The estimated times indicate the overall time required for each step, including reactions or incubations.
Background
Eukaryotic genomic DNA is compactly organized in the nucleus as chromatin, with the nucleosome as its fundamental structural unit [1,2]. The nucleosome consists of approximately 145–147 base pairs of DNA wrapped around a histone octamer, composed of two copies each of histones H2A, H2B, H3, and H4 [3–5]. Structural studies of chromatin have significantly contributed to our understanding of transcriptional regulation, genome stability, and epigenetic inheritance. Biochemical and structural analyses using in vitro–reconstituted chromatin have provided valuable insights into the structural and functional versatility of nucleosomes [6–10]. These reconstituted systems, typically composed of recombinant histones and synthetic DNA templates, offer convenient sample homogeneity and are well-suited for high-resolution structural studies [5,11]. The structures of in vitro–reconstituted nucleosomes are now well characterized, and thus recent efforts have shifted toward understanding their structural and functional properties within the cellular environment, particularly in the context of histone post-translational modifications, native DNA sequences, and inter-nucleosomal interactions.
To better capture chromatin structure within its native cellular context, in situ chromatin analyses using cryo-electron tomography (cryo-ET) have provided significant advancements, aided by sample-thinning techniques such as physical cryosectioning with cryo-microtomes and lamella preparation via cryo-focused ion beam scanning electron microscopy (FIB-SEM) [12–18]. While these approaches are powerful for visualizing chromatin in its native state, they require highly specialized expertise to produce optimal cryo-ET specimens, and access to cryo-FIB-SEM instrumentation remains limited to a small number of research facilities.
To address these limitations, we developed an accessible and robust protocol for preparing native chromatin fragments from HeLa cells for cryo-EM analysis. This method builds on and improves our previously reported chromatin preparation technique [19], combining chemical crosslinking to preserve in situ nucleosome–nucleosome interactions with micrococcal nuclease (MNase) digestion to efficiently solubilize chromatin into mono- and poly-nucleosome fragments. This approach enables high-resolution structural analyses of mono-nucleosomes and provides insights into chromatin organization in a state that closely reflects physiological conditions. Although optimized for HeLa cells, the protocol can be readily adapted to other mammalian cell lines, offering broad applicability for studying chromatin regulation in diverse biological contexts.
Materials and reagents
Biological materials
1. HeLa S3 cells (ATCC, CCL-2.2)
Reagents
1. Dulbecco’s modified Eagle medium (DMEM) (Nacalai, catalog number: 08458-16)
2. Fetal bovine serum (FBS) (Gibco, catalog number: 26140-079)
3. Penicillin-streptomycin mixed solution (Nacalai, catalog number: 09367-34)
4. Dulbecco’s phosphate-buffered saline, liquid [D-PBS(-)] without Ca and Mg (Nacalai, catalog number: 14249-95)
5. 2.5 g/L Trypsin/1 mmol/L EDTA solution (Nacalai, catalog number: 35554-64) (used for cell passaging)
6. NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, catalog number: 78833 or 78835)
7. Protease inhibitor cocktail (EDTA-free) (100×) (Nacalai, catalog number: 03969-34)
8. Formaldehyde solution (36%–38%) (Nacalai, catalog number: 16223-55)
9. Micrococcal nuclease (MNase) (New England Biolabs, catalog number: M0247S)
10. Proteinase K, recombinant, PCR grade (Merck, catalog number: 03115828001)
11. Agarose for 50~800 bp fragment (Nacalai, catalog number: 01147-96)
12. 100 bp DNA ladder (TAKARA, catalog number: 3407A)
13. Ethidium bromide solution (10 mg/mL) (EtBr) (Nacalai, catalog number: 14631-94)
14. Milli-Q water (used directly from the purification system without additional filtration)
15. 2 M glycine (prepared from glycine powder) (Nacalai, catalog number: 17109-35)
16. 1 M Tris-HCl (pH 8.0) (prepared from Tris base) (Nacalai, catalog number: 35434-21)
17. 1 M Tris-HCl (pH 7.5) (prepared from Tris base) (Nacalai, catalog number: 35434-21)
18. 1 M HEPES-NaOH (pH 7.5) (prepared from HEPES) (Nacalai, catalog number: 17514-15)
19. 4 M KCl (prepared from potassium chloride) (Nacalai, catalog number: 28514-75)
20. 5 M NaCl (prepared from sodium chloride) (Nacalai, catalog number: 31320-05)
21. 1 M CaCl2 (prepared from calcium chloride) (Nacalai, catalog number: 08894-25)
22. 1 M DTT (prepared from dithiothreitol powder) (Nacalai, catalog number: 14128-62)
23. 0.5 M EDTA (prepared from EDTA 2Na dihydrate) (Nacalai, catalog number: 15130-95)
24. 10% NP-40 (prepared from Nonidet P40 substitute) (Nacalai, catalog number: 18551-24)
25. 30% sucrose (prepared from sucrose powder) (Nacalai, catalog number: 30403-55)
26. 10% SDS (prepared from sodium lauryl sulfate granular) (Nacalai, catalog number: 02873-75)
27. Acetic acid (Nacalai, catalog number: 00212-43)
28. 1× TAE (prepared from Tris base, acetic acid, and EDTA)
Solutions
1. Buffer A (see Recipes)
2. Proteinase K mix (see Recipes)
3. 10% sucrose gradient solution (see Recipes)
4. 50% sucrose gradient solution (see Recipes)
5. Buffer B (see Recipes)
Recipes
1. Buffer A
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 10 mM | 500 μL |
| 4 M KCl | 200 mM | 2.5 mL |
| 1 M CaCl2 | 1 mM | 50 μL |
| 10% NP-40 | 0.5% | 2.5 mL |
| Milli-Q water | n/a | up to 50 mL |
| Total | n/a | 50 mL |
Used for chromatin fragmentation by MNase. Store at 4 °C.
2. Proteinase K mix
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Proteinase K | 1/2 of the stock | 30 μL |
| 10% SDS (w/v) | 1.67% | 10 μL |
| Milli-Q water | n/a | 20 μL |
| Total | n/a | 60 μL |
Used for digesting proteins in chromatin fragments to estimate DNA size by agarose gel electrophoresis.
3. 10% sucrose gradient solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M HEPES-NaOH (pH 7.5) | 10 mM | 500 μL |
| 5 M NaCl | 30 mM | 300 μL |
| 1 M DTT | 1 mM | 50 μL |
| Sucrose | 10% (w/v) | 5 g |
| Milli-Q water | n/a | up to 50 mL |
| Total | n/a | 50 mL |
Add DTT immediately before use. The lower-density sucrose solution is used for ultracentrifugation.
4. 50% sucrose gradient solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M HEPES-NaOH (pH 7.5) | 10 mM | 500 μL |
| 5 M NaCl | 30 mM | 300 μL |
| 1 M DTT | 1 mM | 50 μL |
| Sucrose | 50% (w/v) | 25 g |
| Milli-Q water | n/a | up to 50 mL |
| Total | n/a | 50 mL |
Add DTT immediately before use. The higher-density sucrose solution is used for ultracentrifugation.
5. Buffer B
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl (pH 7.5) | 10 mM | 5 mL |
| 5 M NaCl | 30 mM | 3 mL |
| 1 M DTT | 1 mM | 500 μL |
| Milli-Q water | n/a | up to 500 mL |
| Total | n/a | 500 mL |
Keep chilled at 4 °C and add DTT immediately before use. Used for buffer exchange after ultracentrifugation prior to cryo-EM sample preparation.
Laboratory supplies
1. Dish for tissue culture (for adhesion cell) 100 mm, 300 pieces (IWAKI, catalog number: 3020-100)
2. Cell scraper (Corning, catalog number: 353085)
3. Micropipette tip (Bioland Scientific, catalog numbers: TIPS10, TIPS200, and TIPS1000)
4. Violamo Centrifuge Tube II (AS ONE Corporation, catalog numbers: VIO-15BN and VIO-50BN)
5. Violamo Disposable Pipette II Easy-Open Package (AS ONE Corporation, catalog numbers: 2-5237-11, 2-5237-03, 2-5237-04, 2-5237-05, and 2-5237-06)
6. 1.5 mL hydrophobic microcentrifuge tube, non-sterile, natural, NoStick (Scientific Specialties Inc., catalog number: 1210-10)
7. 13.2 mL Open-Top Thinwall Ultra-Clear Tube, 14 × 89 mm (Beckman Coulter, catalog number: 344059)
8. Prepacked Disposable PD-10 Columns (Cytiva, catalog number: 17085101)
9. Quantifoil R1.2/1.3 200 Mesh, Cu (Quantifoil, catalog number: M2955C-1)
10. C-Clip Ring (Thermo Fisher Scientific, catalog number: 1036173)
11. C-Clip (Thermo Fisher Scientific, catalog number: 1036171)
Equipment
1. CO2 gas incubator (Astec, model: SCA-80DRS)
2. VP-050N ultrasonic homogenizer (TAITEC, model: 0079435-000)
3. Gradient Master (SK BIO International Co., model: 108)
4. Optima XPN-80 (Beckman Coulter, model: A95765)
5. SW 41 Ti Swinging-Bucket Rotor Package (Beckman Coulter, model: 331336)
6. NanoDrop One (Thermo Fisher Scientific, model: ND-ONE-W)
7. PIB-10 (Vacuum Device Inc.)
8. Vitrobot Mark IV (Thermo Fisher Scientific)
9. Krios G4 cryo-TEM (Thermo Fisher Scientific)
10. K3 BioQuantum direct electron detector (Gatan, model: 1967)
Procedure
文章信息
稿件历史记录
提交日期: Jul 24, 2025
接收日期: Sep 7, 2025
在线发布日期: Sep 17, 2025
出版日期: Oct 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Hatazawa, S., Takizawa, Y. and Kurumizaka, H. (2025). Preparation of Chromatin Fragments From Human Cells for Cryo-EM Analysis. Bio-protocol 15(20): e5472. DOI: 10.21769/BioProtoc.5472.
分类
细胞生物学 > 细胞结构 > 染色体
生物物理学 > 电子冷冻断层扫描
细胞生物学 > 细胞器分离 > 细胞核
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link