- EN - English
- CN - 中文
Fractionation and Extraction of Cell Wall Proteins From Candida albicans
白色念珠菌细胞壁蛋白的分级与提取
(§Deceased) 发布: 2025年09月20日第15卷第18期 DOI: 10.21769/BioProtoc.5456 浏览次数: 2534
评审: Alba BlesaWolf Dieter RötherKyle Stewart Skalenko
Abstract
Candida albicans is the pathogenic fungus that most frequently causes infections in humans. It is part of the microbiota commonly found in the skin, gastrointestinal tract, and vaginal mucosa. However, certain conditions, including immunosuppression, excessive use of antibiotics, hormonal changes, the use of medical devices in patients, and individual nutritional status, promote the development of opportunistic infections caused by this fungus. One of the main fungal structures interacting with the host is the cell wall, which is principally composed of chitin, glucan, and proteins. The cell wall plays key functions for the cell, such as osmotic protection; it is also responsible for cellular shape and acts as a signaling hub in response to environmental changes. Cell wall proteins participate in diverse cellular functions, such as attachment to surfaces and cell wall structure; some possess catalytic or transport activities. In this protocol, we show the methodology for isolating cell wall proteins covalently linked or not to cell wall components that can be previously labeled with [14C]-L-lysine by the action of the fungal transglutaminase localized in the cell wall. We use an extraction method by mechanical cell disruption and washing with 2 M NaCl, whose ionic strength eliminates contaminating proteins from other organelles, through subsequent serial treatments with SDS, chitinase, and zymolyase.
Key features
• Methodology for obtaining cell walls (CW) by mechanical cell disruption, fractionation, and isolation of CW proteins previously labeled with [14C]-L-lysine by the endogenous CW transglutaminase.
• Extraction of CW noncovalently-linked proteins by sequential treatments with 2% SDS, and those covalently-linked CW proteins by chitinase and zymolyase treatment.
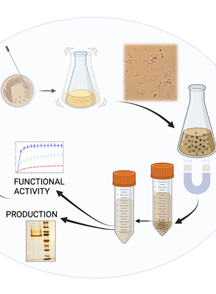
Keywords: Cell fractionation (细胞分级)Graphical overview
Background
Globally, Candida albicans is a pathogenic fungus that frequently affects humans, causing mild skin infections to severe invasive infections. This fungus is part of the normal microbiota of the skin, gastrointestinal tract, and vaginal mucosa. However, several factors, including immunosuppression, excessive use of antibiotics, hormonal changes, usage of medical devices, and individual nutritional status, promote the development of opportunistic infections caused by this fungus. At the cellular level, one of the main structures that has contact with the host is the cell wall, which performs various essential functions for the fungus, such as osmotic protection, signaling center, morphogenesis, and reprogramming of cell wall components in response to the availability of carbon, blood, serum, high temperatures, and environmental acidity [1–3]. The response to these factors promotes changes in the content of cell wall components such as glucan, chitin, and proteins (Figure 1).
The mannosylated protein coat, covalently linked to cell wall polysaccharides, mediates adherence to both the host and medical devices. It also prevents the immune system from recognizing the glucan layer, promotes biofilm formation, and favors communication between the host and C. albicans yeast cells [4].
Given the importance of the C. albicans cell wall proteins, several strategies have been implemented for their extraction and analysis. In a study carried out by our group, cell wall proteins previously labeled with [14C]-L-lysine by the action of the fungal transglutaminase were sequentially extracted with 2% SDS, chitinase, and zymolyase, and identified by tandem mass spectrometry. The zymolyase treatment released 37 proteins, and 41 were identified after incubation with chitinase [1].
The proteins extracted with 2% SDS showed an apparent molecular mass of less than 50 kDa. On the contrary, the proteins released by the zymolyase and chitinase treatments showed a molecular mass greater than 180 kDa. Bioinformatics analysis revealed that the proteins extracted with 2% SDS were reportedly involved in catalytic, binding, structural, and transport functions. The proteins extracted with zymolyase have structural and catalytic functions. In the case of proteins extracted with chitinase, proteins have catalytic, binding, structural, and transport roles. Of the entire universe of identified proteins, 24 were extracted by the three conditions. Moreover, only sixteen have been documented and localized in the cell wall, with Eno1, Pgk1, and Als1 being immunodominant proteins [5–7].
This protocol aims to describe the methodology for extracting cell wall proteins by sequential treatments with 2% SDS to extract noncovalently linked proteins, and extracting those covalently linked to chitin and glucan with chitinase and zymolyase, respectively.
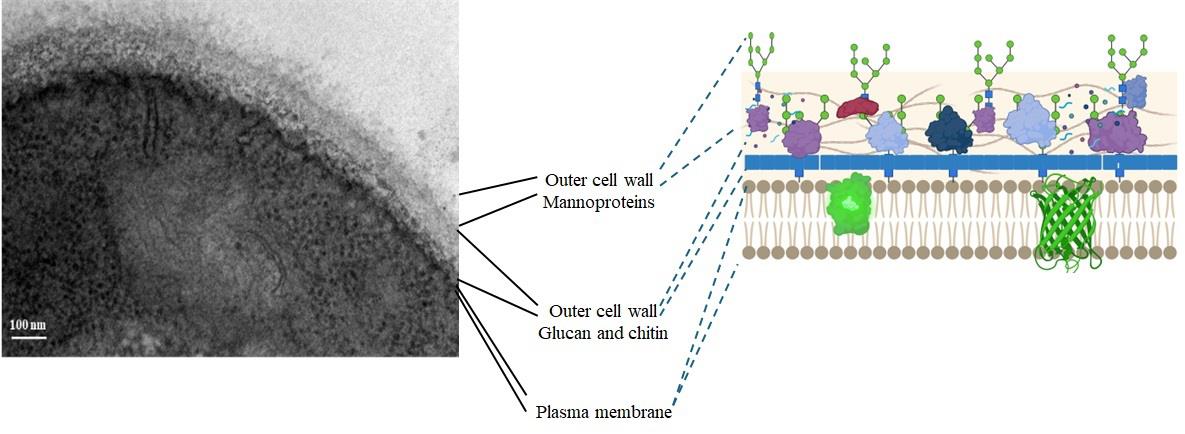
Figure 1. Ultrastructure of the Candida albicans CAI4 yeast cell wall (our results)
Materials and reagents
Biological materials
1. Candida albicans CAI4 strain (Δura3:: λimm434/Δura3:: λimm434) [8]
Reagents
1. Yeast extract (BD, catalog number: 211929)
2. Bacto peptone (MCD LAB, catalog number: 9081)
3. Glucose (Merck, catalog number: 1083371000)
4. Uridine (Sigma-Aldrich, catalog number: U3003-50G)
5. Glass beads, 0.5 mm diameter (BioSpec Products, catalog number: 11079105)
6. CaCl2 (Sigma-Aldrich, catalog number: C4901)
7. Trizma base (Sigma-Aldrich, catalog number: T1503-1KG)
8. HCl (Sigma-Aldrich, catalog number: 258148)
9. Sodium dodecyl sulfate (SDS) (Bio-Rad, catalog number: 161-0302)
10. Zymolyase 20T (MP, catalog number: 0832092)
11. Chitinase from Streptomyces griseus (Sigma-Aldrich, catalog number: C6137)
12. Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T0699-100ML)
13. [14C(U)]-lysine (326 mCi/mL, 3.7 MBq/mL, 50 μCi) Perkin Elmer (NEC-280E), uniformly labeled (all carbon atoms labeled)
14. Phenylmethanesulfonyl fluoride (Sigma-Aldrich, catalog number: P7626)
15. KH2PO4 (Merck, catalog number: 7778-77-0)
16. K2HPO4 (Merck, catalog number: 7758-11-4)
17. NaCl (Sigma-Aldrich, catalog number: S9888)
18. Pepstatin A (Sigma-Aldrich, catalog number: P5318-25MG)
19. Absolute ethanol (J.T. Baker, catalog number: 9000-03)
20. Methanol (J.T. Baker, catalog number: 9070)
21. Developer and replenisher (GBX, Carestream Dental, catalog number: 1900943)
22. Fixer and replenisher (GBX, Carestrean Dental, catalog number: 190 1875)
23. Ready Protein Liquid Scintillation Cocktail (Beckman, catalog number: PN 510814-AA)
24. Coomassie Brilliant Blue R-250 (Bio-Rad, catalog number: 1610400)
25. Isopropyl alcohol (J.T. Baker, catalog number: 49084-02)
26. Acetic acid (J.T. Baker, catalog number: 4908-05)
27. Amplify fluorographic reagent (Amersham, Cytiva, catalog number: 95040-024)
28. N,N-dimethyl casein from bovine milk (Sigma-Aldrich, catalog number: C9801)
29. Distilled deionized water (ddH2O)
Solutions
1. YPD (see Recipes)
2. 50 mM Tris/HCl pH 7.4 (see Recipes)
3. 1 M PMSF (see Recipes)
4. 1 mg/mL pepstatin A (see Recipes)
5. Buffer A (see Recipes)
6. 1 μg/mL pepstatin A (see Recipes)
7. 2 M NaCl (see Recipes)
8. 50 mM phosphate buffer (pH 7.4) (see Recipes)
9. 10 mM CaCl2 (see Recipes)
10. SDS, 2% (w/v) (see Recipes)
11. 5% (v/v) trichloroacetic acid (TCA) (see Recipes)
12. 10% (v/v) TCA (see Recipes)
13. 70% ethanol solution (see Recipes)
14. 75% ethanol solution (see Recipes)
15. Staining solution for protein polyacrylamide gels (see Recipes)
Recipes
1. YPD (1 L)
| Reagent | Final concentration | Quantity or volume |
| Yeast extract | 1% (w/v) | 10 g |
| Bacto peptone | 2% (w/v) | 20 g |
| Glucose | 2% (w/v) | 20 g |
| ddH2O | n/a | 1 L |
Sterilize the uridine stock solution through a sterile 0.22 μm (pore size), 33 mm diameter polyethersulfone filtration device.
Sterilize the YPD medium at 121 °C for 20 min and supplement with 25 μg/mL Uridine, previously sterilized, after cooling the YPD medium to approximately 37–40 °C.
If solid media is required, add 1.5% (w/v) agar to liquid YPD and sterilize by autoclaving at 121 °C for 20 min.
2. 50 mM Tris/HCl pH 7.4 (100 mL)
Mix 0.6057 g of Trizma base in approximately 80 mL of ddH2O. Adjust pH to 7.4 by adding 1 M HCl dropwise. Adjust the volume to 100 mL with ddH2O in a volumetric flask.
3. 1 M PMSF
Dissolve 0.17419 g of phenylmethanesulfonyl fluoride in 1 mL of absolute ethanol and store at -20 °C.
4. 1 mg/mL pepstatin A
Weigh 1 mg of pepstatin A and dissolve in 1 mL of 10% (v/v) acetic acid in methanol.
5. Buffer A (100 mL)
Mix 99.9 mL of 50 mM Tris/HCl pH 7.4 with 0.1 mL of 1 M PMSF. Use immediately.
6. 1 μg/mL pepstatin A
Add 0.1 mL of pepstatin A (1 mg/mL) to 99.9 mL of buffer A.
7. 2 M NaCl
Dissolve 11.688 g of NaCl in 100 mL of ddH2O.
8. 50 mM phosphate buffer (pH 7.4)
Solution A: 13.6 g of KH2PO4 per 1 L (0.1 M) of ddH2O.
Solution B: 17.42 g of K2HPO4 per 1 L (0.1 M) of ddH2O.
Mix 19.0 mL of solution A and 81 mL of solution B for 100 mL of buffer at pH 7.4.
9. 10 mM CaCl2
Dissolve 1.1 mg of CaCl2 per 1 mL of ddH2O.
Add 200 μL to the reaction mixture for labeling proteins with [14C]-L-lysine at 1 mL final volume.
10. SDS, 2% (w/v)
Dissolve 2 g of SDS (sodium dodecyl sulfate) in 100 mL of ddH2O with a low stirring rate to avoid air bubbles. Filter the solution to remove solid contaminants using a 0.22 μm nitrocellulose membrane.
11. 5% (v/v) TCA
Mix 5 mL of 100% TCA solution in 95 mL of ddH2O.
12. 10% (v/v) TCA
Mix 10 mL of 100% TCA solution in 90 mL of ddH2O.
13. 70% ethanol solution
For 100 mL, mix 70 mL of absolute ethanol with 30 mL of ddH2O.
14. 75% ethanol solution
For 100 mL, mix 75 mL of absolute ethanol with 25 mL of ddH2O.
15. Staining solution for protein polyacrylamide gels
For 1 L, dissolve 2 g of Coomassie Brilliant Blue R-250 in a solution of isopropyl alcohol/acetic acid/water (1/1/8, v/v/v).
Laboratory supplies
1. Falcon high-clarity polypropylene 50 mL centrifuge tube (Corning, catalog number: 352070)
2. Microcentrifuge tubes (1.5 mL) (Eppendorf, catalog number: Z606340)
3. Gilson Pipetman pipettes (P1000, P200, P20, P2)
4. Axygen Micropipette tips [1,000 µL (catalog number: T-1000-B), 200 µL (catalog number: T-200-y), 20 µL (catalog number: T-200-y), and 2 µL (catalog number: T-300)]
5. Glass slides (VELAB, catalog number: DBKVEP-20)
6. Glass coverslips (VELAB, catalog number: DBKVE-22)
7. Pyrex Erlenmeyer flasks (100 mL) (Corning, catalog number: CLS4280100)
8. Nitrocellulose filter membrane, pore size 0.45 μm, 47 mm diameter (Millipore, catalog number: N0271)
9. Polyethersulfone filtration device, 0.22 μm (pore size), 33 mm diameter (Millipore, catalog number: LGPM33RS)
10. Open-top thin-wall polypropylene tube, 16 mm × 76 mm (Beckman Coulter, catalog number: 326814)
11. Pyrex Erlenmeyer flasks, 250 mL (Merk, Corning, catalog number: CLS5100250)
12. Whatman glass microfiber grade GF/C filter discs 1.2 μm pore size, 24 mm diameter (Merck, catalog number: WHA1822024)
13. Carestream X-OMAT LS autoradiography film (Sigma-Aldrich, catalog number: F1149)
14. Glass beads 0.5 mm in diameter (BioSpec products, catalog number: 11079105)
15. WHEATON liquid scintillation vial with attached cap (20 mL) (WHEATON, catalog number: DWK986546)
16. 50 mL polypropylene Falcon tubes (Corning, catalog number: CLS352098)
17. Pyrex® VISTA culture tube, rimless, 16 mm diameter × 125 mm in length (Merck, catalog number: CLS7082016X)
18. Cotton swabs (El Crisol, catalog number: LAUKA00904) or Q-tips cotton swabs
19. White Foamboard 5 mm thick, A4 size (Cathedral)
Equipment
1. Water bath incubator (New Brunswick Scientific)
2. Eppendorf 5810R centrifuge
3. Type 90 Ti fixed-angle titanium rotor (Beckman Coulter)
4. NanoDrop 2000 spectrophotometer (Thermo Scientific)
5. Vortex-2 Genie mixer (Sigma-Aldrich)
6. BioSpec BeadBeater homogenizer
7. Epoch 2 ELISA microplate spectrophotometer (BioTek, Agilent Technologies)
8. GelDoc TM EZ imaging system (Bio-Rad)
9. Beckman Coulter LS6500 liquid scintillation counter
10. Ultracentrifuge (Beckman Coulter, model: Optima L-100 XP Class S)
11. Light microscope (Nikon)
12. Analytical balance (Denver Instruments, model: APX-200)
13. Vacuum 1225 Sampling Manifold (Millipore, model: XX2702550)
14. Gel dryer (Bio-Rad, model: 583)
15. Geiger-Müller counter (Dosimeter, model: 3700A)
Software and datasets
1. NanoDrop 2000 software (Thermo ScientificTM, version 1.6)
2. Beckman Coulter LS6500 Scintillation System, using a Motorola 6800 Series microprocessor, a Digital Signal Processor, and a 32,768 channel multichannel analyzer
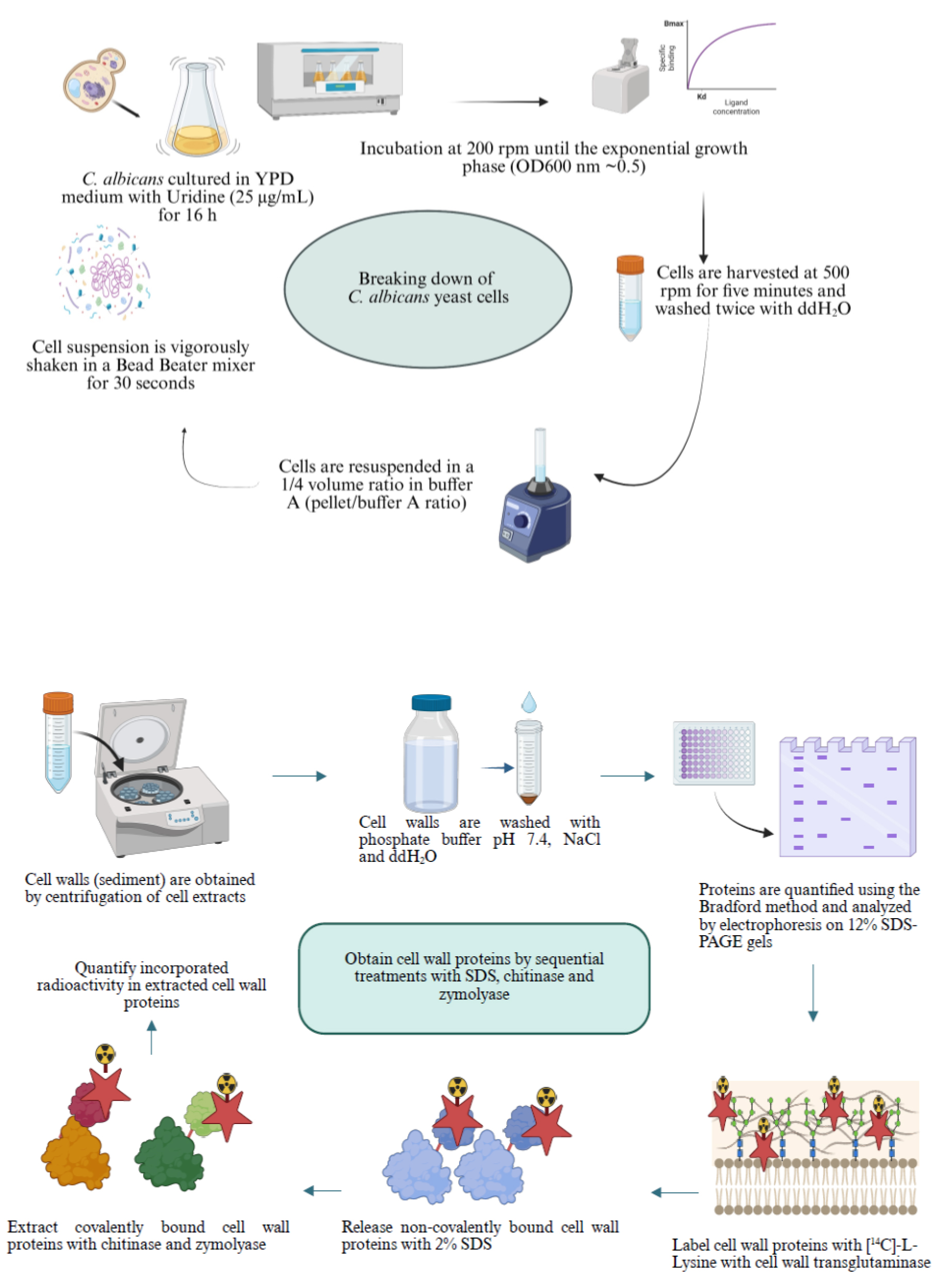
Procedure
文章信息
稿件历史记录
提交日期: Feb 14, 2025
接收日期: Jul 17, 2025
在线发布日期: Sep 2, 2025
出版日期: Sep 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Reyna-Beltrán, E., Iranzo, M., Mormeneo, S., Bazán-Méndez, C. I., Labra-Barrios, M. L., Hernandez-Martínez, E. and Luna-Arias, J. P. (2025). Fractionation and Extraction of Cell Wall Proteins From Candida albicans. Bio-protocol 15(18): e5456. DOI: 10.21769/BioProtoc.5456.
- Reyna-Beltrán, E., Iranzo, M., Calderón-González, K. G., Mondragón-Flores, R., Labra-Barrios, M. L., Mormeneo, S. and Luna-Arias, J. P. (2018). The Candida albicans ENO1 gene encodes a transglutaminase involved in growth, cell division, morphogenesis, and osmotic protection. J Biol Chem. 293(12): 4304–4323. https://doi.org/10.1074/jbc.m117.810440
分类
微生物学 > 微生物细胞生物学 > 细胞器分离
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link