- EN - English
- CN - 中文
Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method
利用等位基因交换方法编辑变形斑沙雷氏菌基因组
发布: 2025年09月20日第15卷第18期 DOI: 10.21769/BioProtoc.5448 浏览次数: 1362
评审: Alba BlesaAnu ThomasAnonymous reviewer(s)
Abstract
No specific ecological niche has been identified for Serratia proteamaculans. Different strains of the bacterium have been described as opportunistic pathogens of plants, animals, and humans, as plant symbionts, and as free-living bacteria. This makes S. proteamaculans and its particular strains promising models for research, particularly aimed at studying the role of various genes in interspecific interactions. Genome editing is one of the most significant approaches used to study gene function. However, as each bacterial species has its own characteristics, editing methods often need to be adapted. In this study, we adapted a conventional approach based on homologous recombination—the allelic exchange method—to edit the genome of S. proteamaculans, with the aim of examining the biological role of protealysin. Plasmids for recombination were created using the suicidal vector pRE118, and then an auxotrophic Escherichia coli ST18 strain was used to deliver these plasmids to S. proteamaculans through conjugation. This method is valid and can potentially be used to create knockouts, knockins, and point mutations in the S. proteamaculans genome, without the need to insert a selective marker into the genome.
Key features
• The genome editing method for Serratia proteamaculans does not require the insertion of selective markers into the genome.
• The selection strategy allows obtaining 30%–40% of clones with the target mutation at the final stage.
• The method can be adapted to introduce knockouts, knockins, and point mutations into the genomes of other bacteria.
Keywords: Serratia proteamaculans (变形斑沙雷氏菌)Graphical overview
Background
Genome editing is a widely used technique when working with bacteria. It is used, in particular, to impart specific properties to microorganisms, search for genes responsible for a particular phenotype, and determine the functions of the studied genes. However, current genome editing methods have been developed for a limited number of bacterial species, and these require adaptation when used with new species. This is due to differences in, for example, cultivation conditions or genomic features.
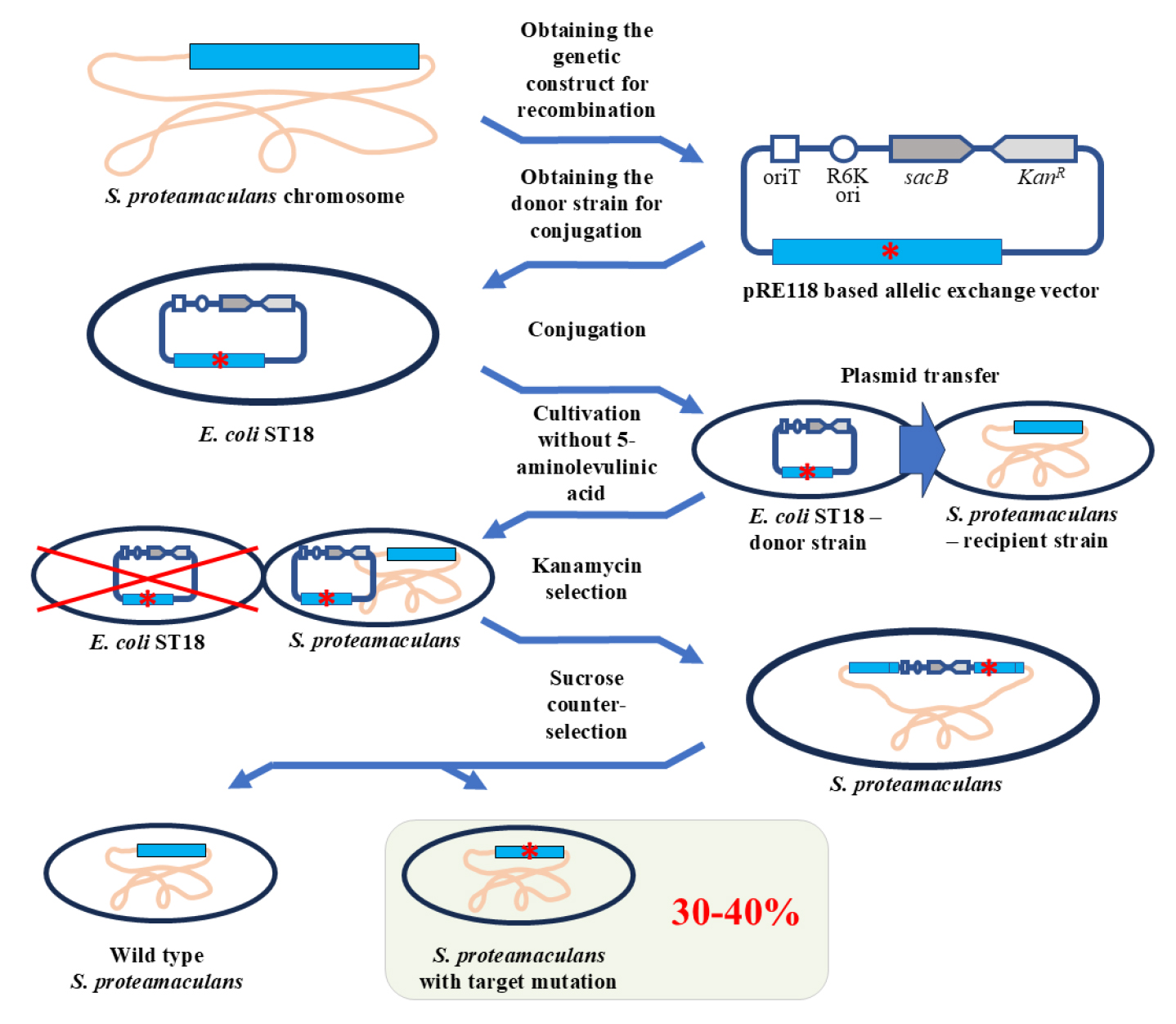
In this protocol, we propose a method for genome editing of Serratia proteamaculans. We developed the protocol to study the biological role of protealysin—a prototype of a new group of proteolytic enzymes, which are widespread among bacteria and are likely involved in pathogenesis [1]. Genome modification methods for S. proteamaculans, the natural producer of protealysin, have not been previously described. Therefore, to edit the S. proteamaculans genome, we adapted a conventional approach based on homologous recombination—the allelic exchange method. The principle of operation of the allelic exchange method is to replace a chromosomal copy with a mutated allele introduced from a plasmid delivered to bacteria. We combined and used the previously described plasmid vector pRE118 [2] and the conjugative E. coli strain ST18 [3]. The vector contains the R6K origin of replication, which only functions in the presence of the pir gene. Since the gene is present in the E. coli ST18 genome, the origin effectively operates in this bacterium, but not in S. proteamaculans. Additionally, pRE118 contains the kanamycin resistance gene for the selection of resistant S. proteamaculans cells in which the plasmid has integrated into the genome through recombination (Figure 1, first crossover). This vector also contains the suicide levansucrase sacB gene, which helps to select cells in which the pRE118 auxiliary elements have been removed from the genome by recombination (Figure 1, second crossover). E. coli ST18 provides plasmid transfer to various recipient bacteria due to the genes from the RP4 plasmid embedded in its chromosome and the transfer origin (oriT) in the pRE118 vector. The advantage of the ST18 strain is that it is auxotrophic for 5-aminolevulinic acid. Thus, the donor strain can be easily removed after conjugation by using a medium without this nutrient, with no need to introduce additional selective markers into the recipient’s genome.
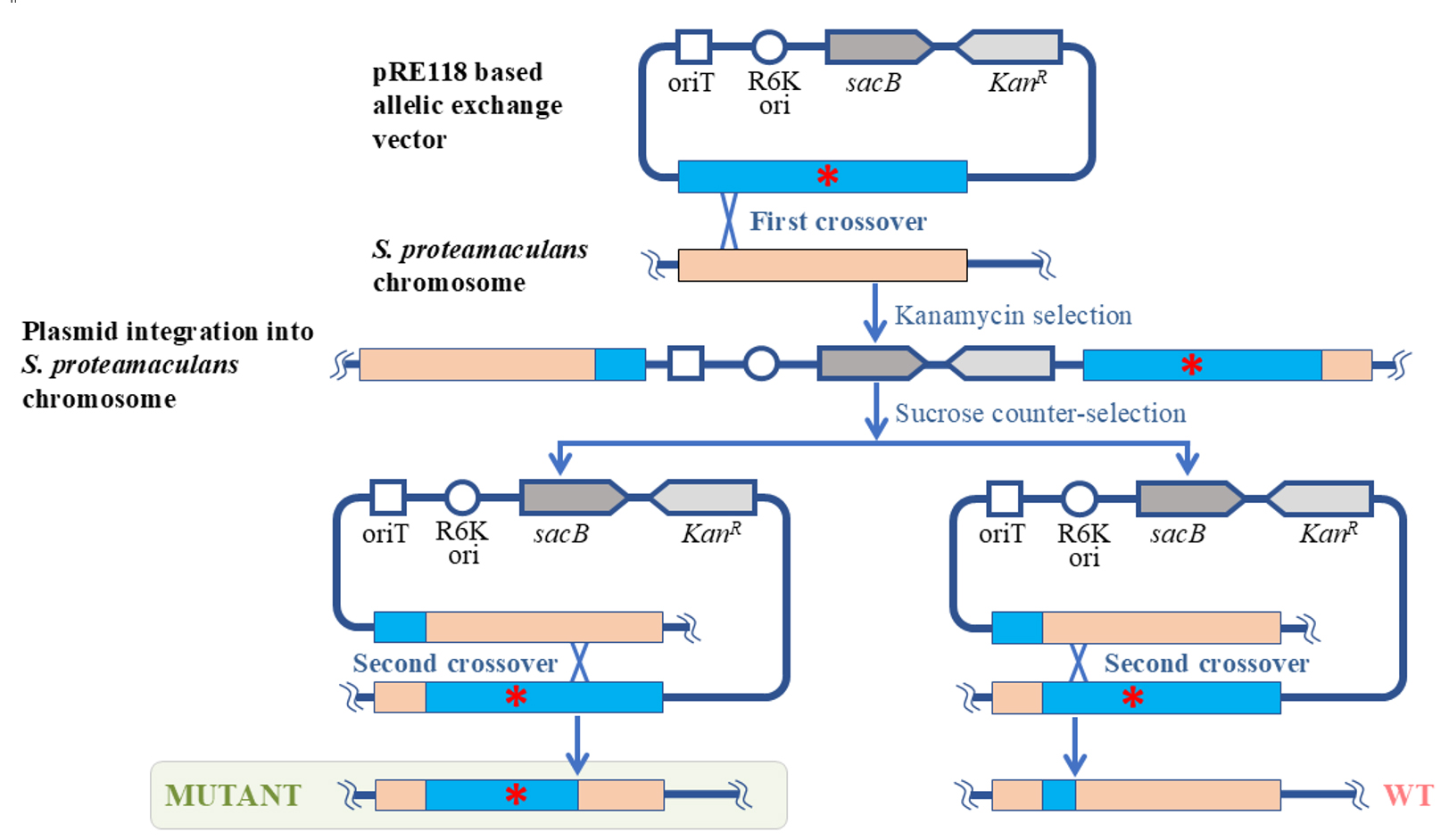
Figure 1. Schematic representation of S. proteamaculans genome editing using the allelic exchange method. The red asterisk denotes any deletion, insertion, or point mutation. oriT: transfer origin; R6K ori: R6K replication origin; sacB: levansucrase gene; KanR: kanamycin resistance gene; WT: wildtype S. proteamaculans.
To delete the part of the S. proteamaculans genome, the homologous recombination cassette (approximately 1,000 bp) was cloned into pRE118. The cassette contained the fragment of genomic DNA with the target deletion. The resulting construct was then introduced using calcium transformation into E. coli ST18 cells, which were further used as donors for plasmid transfer to S. proteamaculans. The first stage of the selection process was carried out on a medium containing kanamycin but no 5-aminolevulinic acid. The second stage took place on a sucrose medium. Sucrose-resistant and kanamycin-sensitive clones were identified and further tested for the target mutations using PCR with the primers specific to the corresponding regions of the bacterial chromosome. Target mutations were detected in 30%–40% of the clones, while the remainder returned to the wild-type allele. The presence of the target mutations was additionally confirmed by sequencing.
We used the described protocol to acquire S. proteamaculans variants with the protealysin gene or protealysin operon deletions [1]. However, this protocol can also be applied to create insertions and point mutations, as was previously described for other bacterial species using a similar approach [4]. Taken together, the proposed protocol allows for successful modification of the S. proteamaculans genome, and it is likely to be applicable to other species from the Serratia genus, as well as other gamma-proteobacteria.
Materials and reagents
Biological materials
1. Strain Serratia proteamaculans 94 [5]
2. Strain Escherichia coli ST18 (DSM 22074) [3]
3. Strain Escherichia coli CC118 (λpir) [6]
4. Plasmid pRE118 (gift from Dieter Schifferli; Addgene plasmid no. 43830) [2]
Reagents
1. Cleanup Standard kit for DNA purification from agarose gels and reaction mixtures (Evrogen, catalog number: BC022S)
2. Plasmid Miniprep kit for plasmid DNA isolation (Evrogen, catalog number: BC221S)
3. Restriction endonucleases XbaI and Psp124BI (an isoschizomer of SacI) and reaction buffer G (SibEnzyme, catalog numbers: SE-E141, SE-E107, and B002)
4. T4 DNA ligase with the reaction buffer (SibEnzyme, catalog number: SE-E329)
5. Q5 High-Fidelity DNA polymerase (New England Biolabs, catalog number: M0491S)
6. Mixture of deoxynucleotide triphosphates (dNTP), 100 mM water solution of each dNTP (SibEnzyme, catalog number: N028)
7. Bacto peptone (Gibco, catalog number: 211820)
8. Yeast extract (Sigma-Aldrich, catalog number: Y1625)
9. Agar powder (BD Difco, catalog number: 281230)
10. Kanamycin sulfate (Gibco, catalog number: 11815024)
11. UltraPure sucrose (Invitrogen, catalog number: 15503022)
12. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888)
13. Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C4901)
14. Phosphate-buffered saline (PBS) tablets (Gibco, catalog number: 18912014)
15. 5-aminolevulinic acid hydrochloride (Sigma-Aldrich, catalog number: A7793)
16. Molecular weight marker DNA ladder (from 0.25 to 10 kb) (SibEnzyme, catalog number: SE-M11)
17. Certified molecular biology agarose (Bio-Rad, catalog number: 1613102)
18. Tris(hydroxymethyl)aminomethane (Merck, catalog number: 1.08387)
19. Boric acid (Sigma-Aldrich, catalog number: 15663)
20. Ethylenediaminetetraacetic acid (Sigma-Aldrich, catalog number: ED)
Solutions
1. Kanamycin stock solution (50 mg/mL) (see Recipes)
2. 5-aminolevulinic acid stock solution (50 mg/mL) (see Recipes)
3. Sucrose stock solution (50% w/w) (see Recipes)
4. 0.1 M CaCl2 solution (see Recipes)
5. PBS buffer (see Recipes)
6. TBE electrophoresis buffer (see Recipes)
7. Agarose 1% (see Recipes)
8. LB medium (see Recipes)
9. Solid LB medium for agar plates (see Recipes)
Recipes
1. Kanamycin stock solution (50 mg/mL)
Dissolve 500 mg of kanamycin sulfate in 10 mL of deionized H2O and store 1 mL aliquots at -20 °C until use.
2. 5-aminolevulinic acid stock solution (50 mg/mL)
Dissolve 50 mg of 5-aminolevulinic acid hydrochloride in 1 mL of deionized H2O, sterilize the solution by filtration through a 0.22 μm membrane, and store at -20 °C until use.
3. Sucrose stock solution (50% w/w)
Dissolve 100 g of sucrose in 100 mL of deionized H2O, sterilize the solution by filtration through a 0.22 μm membrane, and store at 4 °C until use.
4. 0.1 M CaCl2 solution
Weigh 1.11 g of CaCl2 and add deionized H2O to 100 mL, sterilize the solution by filtration through a 0.22 μm membrane, and store at 4 °C until use.
5. PBS buffer
Dissolve one PBS tablet in 100 mL of deionized H2O, sterilize the solution by filtration through a 0.22 μm membrane, and store at 4 °C until use.
6. TBE electrophoresis buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris(hydroxymethyl)aminomethane | 89 mM | 10.8 g |
| Boric acid | 89 mM | 5.5 g |
| Ethylenediaminetetraacetic acid | 2 mM | 0.58 g |
| H2O | n/a | to 1 L |
Dissolve TBE electrophoresis buffer components in deionized H2O and store at room temperature until use.
7. Agarose 1%
Weigh 1 g of agarose and add TBE electrophoresis buffer (see Recipe 6) to 100 mL. Heat the resulting mixture in a boiling water bath until the agarose has completely dissolved and a homogeneous liquid has been obtained. To create the gel, pour into a mold the solution that has been cooled to 45–50 °C.
8. LB medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Peptone | 1% | 10 g |
| Yeast extract | 0.5% | 5 g |
| NaCl | 0.5% | 5 g |
| Kanamycin (optional) | 50 µg/mL | 1 mL of stock solution |
| 5-aminolevulinic acid (optional) | 50 µg/mL | 1 mL of stock solution |
| H2O | n/a | to 1 L |
Dissolve liquid LB medium components in distilled H2O and autoclave the resulting mixture at 121 °C for 45 min. Note that 5-aminolevulinic acid and kanamycin should be added after autoclaving when the medium has been cooled to 40–45 °C.
9. Solid LB medium for agar plates
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Peptone | 1% | 10 g |
| Yeast extract | 0.5% | 5 g |
| NaCl* | 0.5% or 0% | 5 g |
| Agar | 1.5% | 15 g |
| Kanamycin (optional) | 50 µg/mL | 1 mL of stock solution |
| 5-aminolevulinic acid (optional) | 50 µg/mL | 1 mL of stock solution |
| Sucrose (optional)* | 10% | 200 mL of stock solution |
| H2O | n/a | to 1 L |
* If the medium is to be supplemented with sucrose, do not add NaCl.
Add distilled H2O to solid LB medium components and autoclave the resulting mixture at 121 °C for 45 min. Note that sucrose, 5-aminolevulinic acid, and kanamycin should be added after autoclaving when the medium has been cooled to 40–45 °C. Use 20 mL of the medium per 90 × 15 mm Petri plate.
Laboratory supplies
1. Petri dishes, 90 mm × 15 mm (SPL life sciences, catalog number: 10093)
2. Microcentrifuge tubes, 1.5 mL (Biologix, catalog number: 80-1500)
3. PCR tubes, 0.2 mL (Accumax, catalog number: APC-001)
4. Pipette tips (Tarsons, catalog numbers: 521000, 521010, 521020)
5. Cell spreader (SPL life sciences, catalog number: 90050)
6. Centrifuge sterile tube, 15 mL (Accumax, catalog number: ACT-15-R-S)
7. Omnifix Luer Lock Solo syringes, 5 mL (B. Brown)
8. PES sterile syringe filter (0.22 μm) (Alwsci, catalog number: C0000502)
9. Cellulose acetate filters (0.45 μm) (Sartorius, catalog number: 11106-25-N)
10. Syringe filter holders (25 mm) (Sartorius, catalog number: 16517-E)
Note: Sterilize 1.5 mL microcentrifuge tubes and pipette tips by autoclaving at 121 °C for 45 min to work with cell cultures. Assemble the filter holder and membrane filter, wrap the assembled filters in aluminum foil, and sterilize in an autoclave at 121 °C for 45 min. Do not overtighten the filter holder before sterilization to avoid damage to the filter during heating. Tighten it just before transferring the cells to the filter.
Equipment
1. Pipettes (Thermo Fisher Scientific, catalog numbers: 4641010N, 4641040N, 4641070N, 4641100N)
2. Water Purification Systems Proseers (Innova, catalog number: NOVAU)
3. T100 PCR thermal cycler (Bio-Rad, catalog number: 1861096)
4. PowerPac basic power supply (Bio-Rad, catalog number:16450504)
5. Horizontal electrophoresis camera (Helicon, catalog number: SE-2)
6. ChemiDoc EZ imaging system (Bio-Rad, catalog number: 12003153)
7. UV transilluminator, wavelength 302 nm (Analytik Jena US, catalog number: 849-20017-4)
8. Thermostat ThermoMixer C (Eppendorf, catalog number: 5382000015)
9. Water bath (Lab Companion, model name: BW3-05G)
10. High-speed freezing centrifuge (Sigma, model name: 2-16KL)
11. Centrifuge MiniSpin (Eppendorf, catalog number: 022620100)
12. UV-visible absorption spectrometer (Agilent Technologies, model number: 8453) equipped with a fiber-optic ultra-micro measuring cell TrayCell (Hellma, catalog number: 105.800-UVS)
13. Autoclave (Tuttnauer, model type: 2540 ML)
Software and datasets
1. Primer-BLAST [7]
Procedure
文章信息
稿件历史记录
提交日期: Jun 23, 2025
接收日期: Aug 12, 2025
在线发布日期: Aug 28, 2025
出版日期: Sep 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Chukhontseva, K., Karaseva, M. A., Komissarov, A. and Demidyuk, I. (2025). Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method. Bio-protocol 15(18): e5448. DOI: 10.21769/BioProtoc.5448.
分类
微生物学 > 异源表达系统
分子生物学 > DNA > 诱/突变
微生物学 > 微生物遗传学 > 重组
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link