- EN - English
- CN - 中文
Verification of N-Linked Glycosylation of Proteins Isolated from Plant or Mammalian Cell Lines Using PNGase Enzyme
利用 PNGase 酶验证来源于植物或哺乳动物细胞系的蛋白质 N-连接糖基化
(*contributed equally to this work) 发布: 2025年09月20日第15卷第18期 DOI: 10.21769/BioProtoc.5444 浏览次数: 1923
评审: Anonymous reviewer(s)

相关实验方案

Cluster FLISA——用于比较不同细胞系蛋白表达效率及蛋白亚基聚集状态的方法
Sabrina Brockmöller and Lara Maria Molitor
2025年11月05日 1209 阅读

利用基于 Western blot 的生物素化抗体内吞检测法监测膜整合蛋白的内吞过程
Alexandra Graninger and Prasanna Satpute-Krishnan
2025年11月20日 2194 阅读
Abstract
N-glycosylation is a ubiquitous post-translational modification (PTM) that regulates protein folding, stability, and biological function. Accurate identification and validation of N-glycosylation are therefore critical for understanding how glycosylation modulates protein activity. Here, we present a robust workflow for analyzing protein N-glycosylation in both animal and plant systems using peptide-N4-(N-acetyl-β-glucosaminyl) asparagine-amidase A and F (PNGase A and PNGase F). After enzymatic cleavage of the asparagine-linked N-glycans, samples are analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting (WB) to detect shifts in apparent molecular weight (MW) indicative of deglycosylation. Key steps include denaturing the protein to expose glycosylation sites, optimizing buffer conditions for PNGase F and A treatment, and comparing glycosylated vs. deglycosylated forms by electrophoretic mobility. A troubleshooting guide addresses common challenges, including incomplete deglycosylation and low transfer efficiency during WB, offering practical solutions to ensure reliable results. This protocol provides researchers with a standardized, cost-effective framework for investigating protein N-glycosylation in diverse systems, from cell lysates to purified proteins, in both animal and plant models.
Key features
• This method employs standard biochemical techniques, such as enzymatic digestion and SDS-PAGE, making it highly accessible and relatively quick to perform.
• Successful deglycosylation results in a detectable downward shift in protein migration on an SDS-PAGE gel. This shift provides a visually confirmable indication of N-glycan removal.
• Prior to engaging in mass spectrometry analyses, this approach serves as a rapid, cost-effective preliminary screening or validation tool for assessing N-glycosylation status.
• Broad system compatibility: PNGase F and A enzymes are active in mammalian, plant, and microbial systems, allowing reliable N-glycosylation assessment regardless of sample origin.
Keywords: Enzymatic assay (酶学检测)Graphical overview
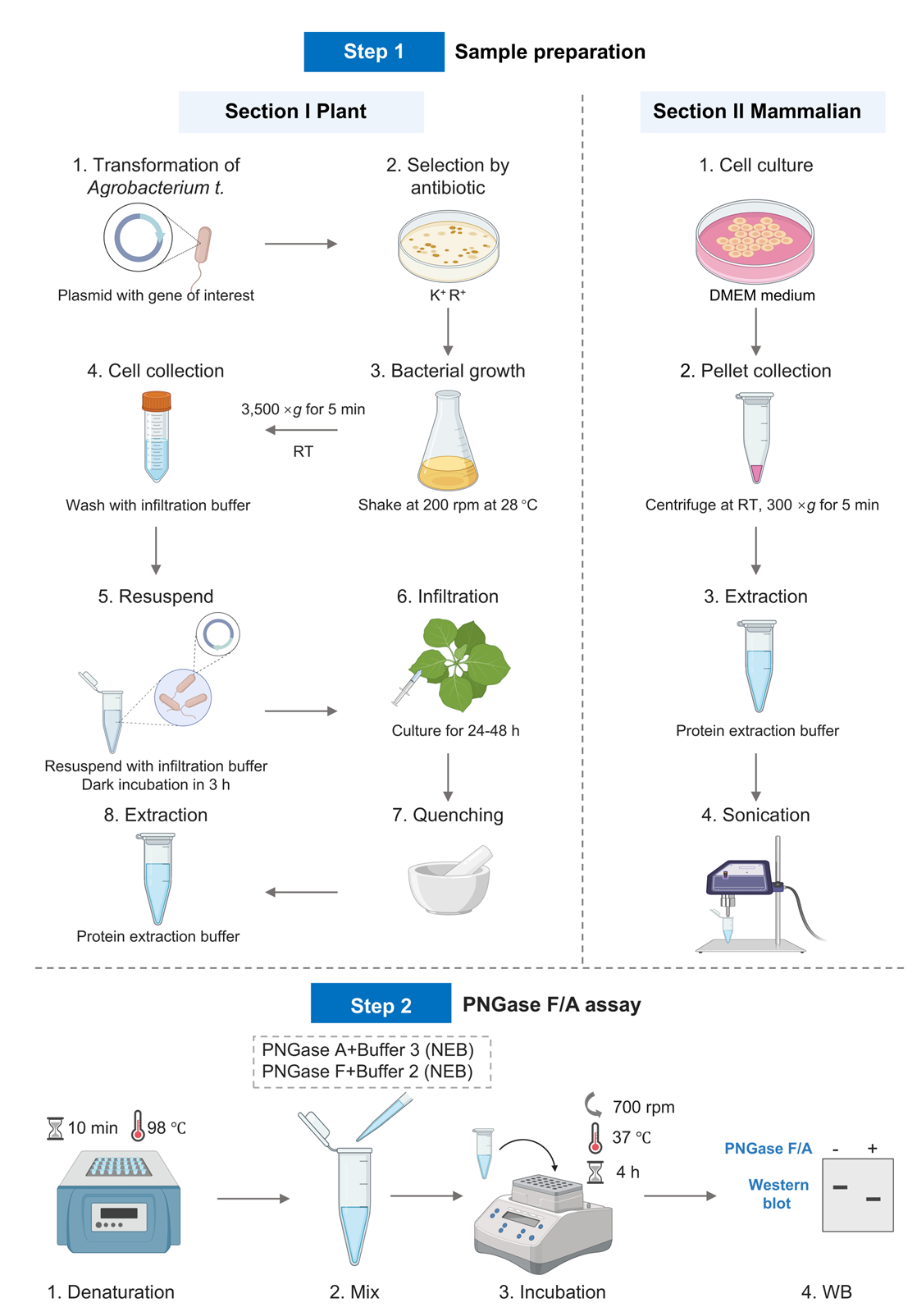
Background
N-linked glycosylation is a major post-translational modification (PTM) in nature, where oligosaccharide chains covalently attach to the amide nitrogen of asparagine (Asn) residues within the classic Asn-X-Ser/Thr (X ≠ Pro) sequence in nascent polypeptides [1]. This modification regulates protein folding, glycan-dependent quality control processes in the endoplasmic reticulum (ER), protein stability, and protein–protein interactions [2]. N-glycosylation is a ubiquitous PTM, with at least 2,000 N-glycosylation sites identified in model organisms such as Arabidopsis thaliana, Drosophila melanogaster, and zebrafish [3], though this number has likely been underestimated.
N-glycosylation plays a crucial role in protein function, trafficking, and immune responses in plants. In Arabidopsis, mutation of N-glycosylation sites in KORRIGAN1 (KOR1) disrupts proper protein folding, Golgi trafficking, and root development [4]. N-glycosylation also regulates the localization of SnRK2.2/2.3 and the brassinosteroid receptor BRI1 [5, 6] and modulates ligand binding of vacuolar sorting receptors such as AtVSR1 [7]. In the EFR receptor, mutation of asparagine at position 143 (Asn143) abolishes ligand-binding capacity, thereby impairing immune responses [8]. In the MIK2 receptor, N-glycosylation at Asn 410 is critical for its interaction with BAK1 [9,10]. In rice, N-glycosylation is essential for the function of the ER-resident lectin OS9 in ER-associated degradation (ERAD) [11]. In wheat, N-glycosylation is pivotal for TaCERK-mediated defense against wheat yellow mosaic virus infection [12]. Despite the identification of thousands of N-glycosylated proteins through omics approaches, functional studies of N-glycosylation on individual plant proteins remain in the early stages, largely due to the limited availability of robust methods for validating N-glycan modifications.
The initial steps of N-glycosylation are highly conserved between plants and animals [2]. However, due to the presence of unique modifications in the plant-specific N-glycosylation pathway, particularly the incorporation of β1,2-xylose and α1,3-fucose residues into the core structure, there are distinct differences in the verification of N-glycan modifications on plant vs. animal proteins [13]. PNGase F, an amidase, can hydrolyze the β-aspartyl glycosylamine bond between the innermost N-acetylglucosamine (GlcNAc) and asparagine, releasing the intact oligosaccharide and converting the Asn to aspartic acid [14]. Unlike endoglycosidases (e.g., Endo H) that cleave within the core glycan, PNGase F can remove all N-linked glycans, making it the gold standard for complete deglycosylation [15]. Its activity requires the protein to be denatured to expose glycosylation sites that may be obscured by the tertiary structure. This enzyme is indispensable in studies where N-glycosylation needs to be precisely validated, such as distinguishing glycosylation isoforms in SDS-PAGE or mapping glycosylation sites by mass spectrometry. PNGase F removes all complex, hybrid, and high-mannose-type glycans and is widely used for N-glycan cleavage in mammalian systems [16]. However, its activity is limited by the presence of α1,3-core fucose in the glycan, which is common in plant systems. In contrast, PNGase A, derived from Oryza sativa, can hydrolyze N-glycans on glycoproteins/peptides regardless of the presence of xylose or fucose (α1-3 or α1-6) (Table 1) [17]. This renders PNGase A the gold standard for complete removal of N-glycans from plant-derived proteins.
Table 1. PNGase F and PNGase A
| Enzyme | PNGase F | PNGase A |
|---|---|---|
| Source | Elizabethkingia spp | Oryza sativa |
| Core specificity | High-mannose-type, complex-type, and hybrid-type without α1-3 core fucosylated glycans | Cleaves α1-3 and α1-6 core fucosylated and high-mannose-type glycans |
| Optimal pH | 7.5 | 4.5–5.5 |
| Compatibility | Mammalian system | Mammalian, plant, and insect systems |
In addition to PNGase F and A, various other enzymes target specific glycan structures or linkage types. For example, endoglycosidases (Endo H/F/S) cleave within the core glycan, leaving a single GlcNAc residue [18]. A comparative overview of key glycan-cleaving enzymes is provided in Table 2, sourced from [18] and the NEB website. In some cases, sequential digestion with enzyme combinations (e.g., sialidase + PNGase F) can enhance the efficiency of glycan removal from highly modified proteins. In this study, a detailed protocol has been established, with PNGase F and PNGase A as the core analytical tools. Specifically, the N-glycosylation of exogenously transformed plant proteins is detected using the plant system, while the N-glycosylation of endogenous animal proteins is examined via the animal system.
Table 2. Enzymes for glycan cleavage
| Enzyme | Source | Substrate specificity | Optimal pH | Key applications |
| Endo H | Streptomyces plicatus | Cleaves high-mannose and hybrid N-glycans | 5.5 | ER-resident glycoprotein analysis |
| Endo F1/F2/F3 | Elizabethkingia spp. | F1: High mannose | F1: 5.5 | Glycoengineering antibody glycosylation patterns |
| F2: Bi-antennary | F2: 4–6 | |||
| F3: Tri-antennary | F3: 5.5 | |||
| Endo S | Streptococcus pyogenes | Cleaves N-glycans on IgG Fc region | 5.5 | Antibody glycan profiling without full digestion |
| O-Glycosidase | Enterococcus faecalis | Removes core 1 O-linked glycans (Galβ1-3GalNAc) | 5.5–7.0 | O-glycan validation (e.g., mucins) |
| Sialidase | Clostridium perfringens | Hydrolyzes α2-3/6/8/9-linked sialic acids | 5.0–6.0 | Desialylation for glycan accessibility studies |
Materials and reagents
Biological materials
1. Nicotiana benthamiana
2. Agrobacterium tumefaciens strain GV3101
3. HEK 293T cell line
Reagents
1. Plant expression vector pCAMBIA2301
2. Rifampicin (Rif) (Sigma, catalog number: R3501)
3. Kanamycin (Kana) (Sigma, catalog number: 420311)
4. DMEM (Gibco, catalog number: 11995065)
5. FBS (Gibco, catalog number: A5256701)
6. Penicillin/streptomycin (Gibco, catalog number: 15140122)
7. Trypsin-EDTA (Gibco, catalog number: 25200056)
8. MES (Sigma, catalog number: M8250)
9. MgCl2 6H2O (Sigma, catalog number: M2393)
10. Acetosyringone (AS) (Sigma, catalog number: D134406)
11. PNGase F (New England Biolabs, catalog number: P0705S)
12. PNGase A (New England Biolabs, catalog number: P0707S)
13. Sodium dodecyl sulfate (SDS) (Sigma, catalog number: L4390)
14. Sodium chloride (NaCl) (Sigma, catalog number: S7653)
15. Potassium chloride (KCl) (Sigma, catalog number: P5405)
16. Disodium hydrogen phosphate (Na2HPO4·12H2O) (Sigma, catalog number: 106579)
17. Potassium dihydrogen phosphate (KH2PO4) (Sigma, catalog number: 104873)
18. Dithiothreitol (DTT) (Sigma, catalog number: D0632)
19. Nonidet-40 (NP-40) (Sigma, catalog number: I8896)
20. Methanol (Sigma, catalog number: 34860)
21. Chloroform (Sigma, catalog number: 319988)
22. 4%–20% gradient SDS-PAGE gel (LabLead, catalog number: P42012)
23. Protein ladder (Thermo Scientific, catalog number: 26616)
24. 5× loading buffer (LabLead, catalog number: G2527)
25. Tris (Aladdin, catalog number: T105287)
26. Glycine (Aladdin, catalog number: G432934)
27. BSA (Aladdin, catalog number: B265993)
28. Primary antibodies: Anti-SEL1L (ABclonal, catalog number: A12073), anti-Myc (MBL, catalog number: M047-3), anti-actin for plants (Abmart, catalog number: M20009), and anti-actin for animals (Beyotime, catalog number: AF0003)
29. Secondary antibodies: HRP-conjugated anti-mouse IgG (Cell Signaling, catalog number: 7076S) and HRP-conjugated anti-rabbit IgG (Cell Signaling, catalog number: 7074S)
30. Liquid nitrogen
31. 10% glycerol (Coolaber, catalog number: SL9280)
32. ECL substrate (Millipore, catalog number: WBKLS0050)
33. Tryptone (Aladdin, catalog number: T139519)
34. Yeast extract (Aladdin, catalog number: Y278690)
Solutions
1. Antibiotic stock solutions (see Recipes)
2. LB liquid medium (see Recipes)
3. Induction buffer (see Recipes)
4. Infiltration buffer (see Recipes)
5. 1× PBS (see Recipes)
6. Protein extraction buffer (see Recipes)
7. Denaturing buffer (1 M DTT) (see Recipes)
8. 10% NP-40 (see Recipes)
9. Transfer buffer (see Recipes)
10. Cell culture medium (see Recipes)
Recipes
1. Antibiotic stock solutions
| Antibiotic | Stock concentration | Quantity (per mL) | Storage |
|---|---|---|---|
| Rif | 50 mg/mL (liquid) | 50 mg | -20 °C, protected from light |
| Kana | 50 mg/mL (liquid) | 50 mg | -20 °C |
2. LB liquid medium
| Reagent | Final concentration | Quality or volume (per L) |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| NaCl | 10 g/L | 10 g |
| Ultrapure water (e.g., Milli-Q) | 1,000 mL |
After autoclaving and cooling to approximately 50 °C, add rifampicin [final 50 μg/mL (1:1,000 dilution from 50 mg/mL stock)] and kanamycin [final 50 μg/mL (1:1,000 dilution from 50 mg/mL stock)].
3. Induction buffer
| Reagent | Final concentration | Quality or volume (per L) |
|---|---|---|
| MES (pH 5.7) | 10 mM | 1.9524 g |
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| NaCl | 10 g/L | 10 g |
| AS | 20 μM | 3.924 mg |
| Ultrapure water (e.g., Milli-Q) | 1,000 mL |
a. Prepare 500 mL of 2× LB medium, then add 400 mL of water.
b. Perform autoclave sterilization and cool to room temperature.
c. Add 100 mL of 0.1 M sterile MES (pH 5.7). Store the prepared medium at 4 °C.
d. Add AS before use to achieve a final concentration of 20 μM.
e. Add Rif and Kana (see Recipe 1) to a final concentration of 50 μg/mL (1:1,000 dilution from 50 mg/mL stock).
4. Infiltration buffer
| Reagent | Final concentration | Quality or volume (per L) |
|---|---|---|
| MES (pH 5.7) | 10 mM | 1.9524 g |
| MgCl2 6H2O | 10 mM | 2.03 g |
| AS | 150 μM | 29.4 mg |
| Ultrapure water (e.g., Milli-Q) | 1000 mL |
a. Dissolve MES and MgCl2 in 800 mL of H2O and adjust pH to 5.7 with KOH.
b. Add AS (dissolved in DMSO) and bring to 1 L. Filter-sterilize and use fresh.
5. 1× PBS
| Reagent | Final concentration | Quality (per L) |
|---|---|---|
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| Na2HPO4·12H2O | 10 mM | 3.58 g |
| KH2PO4 | 1.8 mM | 0.24 g |
a. Dissolve all reagents in 800 mL of ultrapure water.
b. Adjust pH to 7.4 with HCl (if needed).
c. Bring the final volume to 1 L with ultrapure water.
d. Sterilize by autoclaving (121 °C, 20 min) or filter sterilization (0.22 μm).
6. Protein extraction buffer
| Reagent | Final concentration | Quality or volume (per 100 mL) |
|---|---|---|
| SDS | 4% (w/v) | 4 g |
| 1× PBS (Recipe 5) | 1× | 100 mL (adjust final volume) |
| Triton | 1% (w/v) | 1 mL |
7. Denaturing buffer (1 M DTT)
| Reagent | Final concentration | Quantity or volume (per 1 mL) |
|---|---|---|
| DTT | 1 M | 0.15425 g |
| Ultrapure water (e.g., Milli-Q) | Up to 1 mL |
Store aliquots at -20 °C. Avoid freeze-thaw cycles.
8. 10% NP-40
| Reagent | Final concentration | Quantity or volume (per 100 mL) |
|---|---|---|
| NP-40 | 10% (v/v) | 10 mL |
| Ultrapure water (e.g., Milli-Q) | 90 mL (adjust to final volume) |
a. Use a graduated cylinder or pipette to measure 10 mL of NP-40 (wear gloves and eye protection; NP-40 is viscous and irritant).
b. Add the NP-40 to approximately 80 mL of ultrapure water in a beaker or bottle.
c. Stir gently with a magnetic stirrer or shake until fully homogeneous (NP-40 dissolves easily but is viscous).
d. Bring the final volume to 100 mL with ultrapure water.
e. Filter through a 0.22 μm filter.
f. Store protected from light.
9. 10× transfer buffer
| Reagent | Final concentration | Quality or volume (per 1,000 mL) |
|---|---|---|
| Tris base | 250 mM | 30.3 g |
| Glycine | 1.92 M | 144 g |
| dH2O | 1,000 mL |
10. Cell culture medium
| Reagent | Final concentration | Volume (per 500 mL) |
|---|---|---|
| DMEM | 500 mL | |
| FBS | 10% | 50 mL |
| Penicillin/streptomycin | 1% | 5 mL |
Store at 4 °C.
Equipment
1. Metal bath (Thermo Scientific, catalog number: 88870001)
2. Sonicator (SCIENTZ-IID, catalog number: JY92-IIN)
3. Thermal mixer (Eppendorf, catalog number: 5382000074)
4. SDS-PAGE electrophoresis system (Bio-Rad, catalog number: 1645070 and 1658001)
5. Wet transfer apparatus (Bio-Rad, catalog number: 1703935)
6. Imaging system (Tanon Chemi Dog Ultra)
Software and datasets
1. ImageLab (Bio-Rad) for gel image analysis
2. UniProt: Reference molecular weights of target proteins
3. BioRender.com online tool: Graphical overview
Procedure
文章信息
稿件历史记录
提交日期: Jun 29, 2025
接收日期: Aug 11, 2025
在线发布日期: Aug 22, 2025
出版日期: Sep 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Hong, W., Lei, C., Qiu, Y., Zhou, Y., Hu, Y., Chen, X., Li, X. and Li, J. (2025). Verification of N-Linked Glycosylation of Proteins Isolated from Plant or Mammalian Cell Lines Using PNGase Enzyme. Bio-protocol 15(18): e5444. DOI: 10.21769/BioProtoc.5444.
分类
生物化学 > 蛋白质 > 修饰
生物化学 > 蛋白质 > 免疫检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









