- EN - English
- CN - 中文
New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
检测和分离拟南芥种子黏液中鼠李半乳糖醛酸Ⅱ的新方法
发布: 2025年09月05日第15卷第17期 DOI: 10.21769/BioProtoc.5433 浏览次数: 1248
评审: Ying LiAnonymous reviewer(s)
Abstract
Rhamnogalacturonan-II (RG-II) is one of the least studied domains of pectin, primarily due to its low abundance, the lack of reliable antibodies, and the complexity of its structure. The present study builds upon existing protocols and procedures used to analyse RG-II in tissues where it is more abundant, combining and adapting them for the isolation of RG-II from Arabidopsis seed mucilage—a structure previously thought to lack RG-II. By applying these adapted methods, we first confirmed the presence of RG-II in seed mucilage and subsequently succeeded in isolating it from a tissue where it is typically present in low abundance, thereby enabling future studies on this previously overlooked component.
Key features
• An efficient RG-II isolation protocol offers the opportunity to simplify and deepen the study of RG-II.
• It provides reliable yields.
• The protocol allows analysis of RG-II synthesis or structural changes affecting dimerisation in mutant lines using Arabidopsis seed mucilage.
Keywords: Rhamnogalacturonan-II (鼠李半乳糖醛酸Ⅱ)Graphical overview
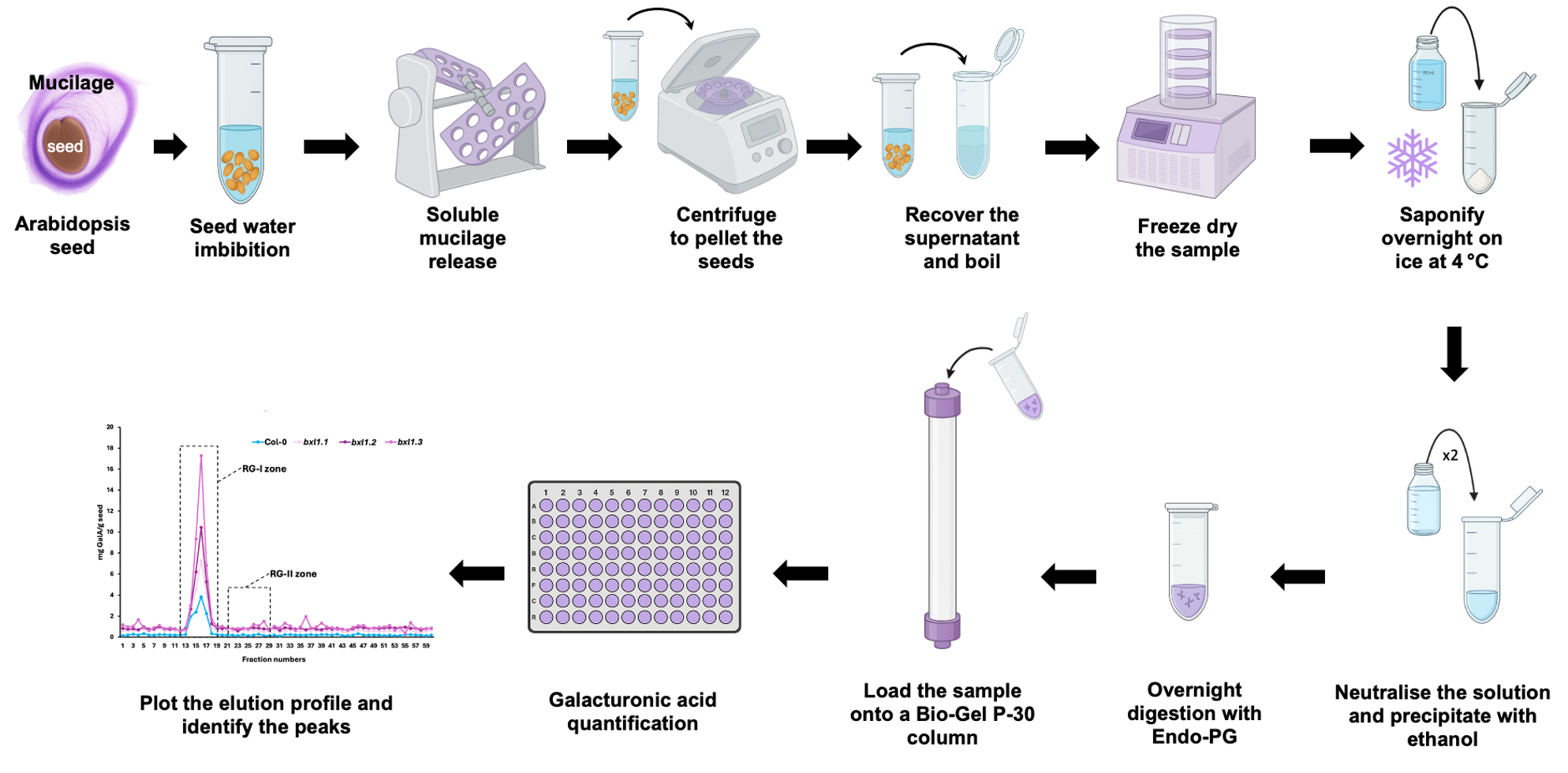
Background
The mucilage of the Arabidopsis seed coat is a gel-like structure produced by specialised epidermal cells [1]. It contains all major polysaccharide groups found in plant cell walls, with pectins comprising 90%–95% of its composition, alongside smaller amounts of other components. The mucilage is predominantly composed of rhamnogalacturonan-I (RG-I), with lesser quantities of homogalacturonan (HG), cellulose, galactoglucomannans, xylans, xyloglucans, and arabinogalactan proteins (AGPs) [2–5]. Although rhamnogalacturonan-II (RG-II) has been proposed to contribute to mucilage organisation, its presence within the mucilage layers remains largely unexplored [5]. Research on RG-II has often been limited due to its low abundance and the absence of reliable analytical tools. Unlike what occurs with plant tissues, where the amount of material is greater and RG-II is more abundant compared to mucilage, it is not possible in this case to directly visualise RG-II without prior enrichment of this domain. In this report, isolated mucilage was processed to evaluate the feasibility of applying protocols previously developed for RG-II-rich tissues [6] to detect and isolate this domain from Arabidopsis seed mucilage. We optimised the amount of dry seed required to extract and process the mucilage needed for size-exclusion chromatography, and successfully isolated RG-II despite its low abundance. By taking advantage of the distinct sizes of RG-I and RG-II following enzymatic digestion with endo-polygalacturonase, we were able to specifically isolate RG-II by determining the elution profile through galacturonic acid quantification [7]. To confirm that the isolated domain was indeed RG-II, we monomerised the dimeric form and conducted a dimerisation assay [6], validating the identity of the recovered polymer. This approach provides a valuable tool for investigating polysaccharide biosynthesis—particularly that of RG-II—using mutants with defects in mucilage structure or function.
Materials and reagents
Biological materials
1. Seeds of Arabidopsis thaliana Col-0
2. Seeds of Arabidopsis thaliana bxl1 (mutant)
Reagents
1. MilliQ water
2. Pyridine (Sigma-Aldrich, CAS: 110-86-1)
3. Acetic acid, glacial (Merck, CAS: 64-19-7)
4. Endo-polygalacturonase (Megazyme, CAS: 9032-75-1)
5. Ammonium persulfate (Thermo Scientific, CAS: 7727-54-0)
6. Tris-base (Merck, CAS: 77-86-1)
7. Hydrochloric acid (HCl) (Merck, CAS: 7647-01-0)
8. Tetramethylethylenediamine (TEMED) (Thermo Scientific, catalog number: 17919)
9. Ethanol (Merck, CAS: 64-17-5)
10. Sodium thiosulphate (Sigma-Aldrich, CAS: 7772-98-7)
11. Silver nitrate (Merck, CAS: 7761-88-8)
12. Formaldehyde (Winkler, CAS: 50-00-0)
13. Sodium carbonate (Na2CO3) (Sigma-Aldrich, CAS: 497-19-8)
14. Sulfuric acid (Merck, CAS: 7664-93-9)
15. Sodium hydroxide (NaOH) (Sigma-Aldrich, CAS: 1310-73-2)
16. 3-Hydroxybiphenyl (Sigma-Aldrich, CAS: 580-51-8)
17. Chlorobutanol (Sigma-Aldrich, CAS: 6001-64-5)
18. Sodium tetraborate decahydrate (Borax decahydrate) (Sigma-Aldrich, CAS: 1303-96-4)
19. D-(+)-Galacturonic acid monohydrate (GalA) (Merck, CAS: 91510-62-2)
20. Lead nitrate (Merck, CAS: 10099-74-8)
21. Glycine (Merck, CAS: 56-40-06)
22. Acrylamide/Bis acrylamide (29:1) 40% (Bio-Rad, catalog number 1610147)
23. Bromophenol blue (Merck, CAS: 115-39-9)
Solutions
1. 0.5 M Na2CO3 (see Recipes)
2. Pyridine:acetic acid:water (PyAW, 1:10:200 v/v/v), 0.25% chlorobutanol (see Recipes)
3. Pyridine:acetic acid:water (PyAW, 1:1:98 v/v/v), 0.25% chlorobutanol (see Recipes)
4. Solution A (see Recipes)
5. Solution B (see Recipes)
6. Galacturonic acid stock solution (see Recipes)
7. Tris-HCl 1.5 M, pH 8.8 (see Recipes)
8. Ammonium persulfate 10% (APS) (see Recipes)
9. Polyacrylamide gel 26.4% (see Recipes)
10. Running buffer for PAGE (see Recipes)
11. Loading buffer (see Recipes)
12. Fixing solution (see Recipes)
13. 400 µM sodium thiosulfate solution (see Recipes)
14. 6 mM silver nitrate and 10 mM formaldehyde solution (see Recipes)
15. Developing solution (see Recipes)
16. Stopping solution (see Recipes)
Recipes
1. 0.5 M Na2CO3
Add 5.3 g of Na2CO3 to 100 mL of MilliQ water.
2. Pyridine:acetic acid:water (PyAW, 1:10:200 v/v/v), 0.25% chlorobutanol
Example for 100 mL:
| Reagent | Quantity/Volume |
|---|---|
| Pyridine | 47 μL |
| Acetic acid | 4.74 mL |
| Water | 9.47 mL |
| Chlorobutanol | 0.25 g |
3. Pyridine:acetic acid:water (PyAW, 1:1:98 v/v/v), 0.25% chlorobutanol
Example for 1 L:
| Reagent | Quantity/Volume |
|---|---|
| Pyridine | 10 mL |
| Acetic acid | 10 mL |
| Water | 980 mL |
| Chlorobutanol | 2.5 g |
4. Solution A
Add 0.5 g of Borax decahydrate to 100 mL of sulfuric acid.
5. Solution B
Add 0.4 g of NaOH to 10 mL of water; then, add 0.015 g of 3-hydroxybiphenyl.
6. Galacturonic acid stock solution
Prepare a 0.1 mg/mL stock solution of galacturonic acid (GalA) by dissolving 1 mg of GalA in 10 mL of MilliQ water.
7. Tris-HCl 1.5 M, pH 8.8
To prepare 1 L, weigh 181.65 g of Tris base and dissolve it in 700 mL of MilliQ water. Add 31 mL of hydrochloric acid, and then adjust the pH to the desired value using 1 M HCl. Finally, bring the volume up to 1 L with MilliQ water.
8. Ammonium persulfate 10% (APS)
To prepare 10 mL, add 1 g of ammonium persulfate to 10 mL of MilliQ water.
9. Polyacrylamide gel 26.4%
Example for 5 mL (one mini gel, using 0.75 mm spacers):
| Reagent | Quantity |
|---|---|
| Tris base 1.5 M pH 8.8 | 834 μL |
| Acrylamide/Bis acrylamide (29:1) 40% | 3.33 mL |
| APS | 46.7 μL |
| TEMED | 3.9 μL |
| Water | 834 μL |
10. Running buffer for PAGE
Example for 1 L (10×):
| Reagent | Quantity |
|---|---|
| Tris base | 60.57 g |
| Glycine | 28.53 g |
11. Loading buffer
To prepare 20 mL of buffer, dissolve 1.53 g of Tris in 7 mL of water, adjust the pH to 6.8 with 1 M HCl, and make up to 10 mL with water. Then, add 0.05 g of bromophenol blue and 10 mL of glycerol.
12. Fixing solution
Example for 1 L:
| Reagent | Quantity/Volume |
|---|---|
| Ethanol | 400 mL |
| Acetic acid | 100 mL |
| Water | 500 mL |
13. 400 µM solution of sodium thiosulfate
Add 63.2 mg of sodium thiosulfate to 1 L water.
14. 6 mM silver nitrate and 10 mM formaldehyde solution
Add 0.102 g of silver nitrate and 75.2 μL of 37% formaldehyde to 100 mL of water.
Note: This solution must be freshly prepared.
15. Developing solution
Add 29.68 g of sodium carbonate, 20 mL of 400 µM sodium thiosulfate solution (Recipe 13), and 4.8 mL of 37% formaldehyde to 975.2 mL of water.
16. Stopping solution
Add 40 g of Tris to 980 mL of water, then complete to 1 L with 20 mL of glacial acetic acid.
Laboratory supplies
1. Bio-Gel P-30 (Bio-Rad, catalog number: 1504154)
2. 96-well plate (Nest, catalog number: 514201)
3. Epoch 96-well plate absorbance reader (Agilent BioTek Epoch Microplate Spectrophotometer, catalog number: 11-120-572)
Equipment
1. Pipettes (Corning, model: LambdaTM Plus Single, catalog numbers: 4070, 4082, and 4075)
2. Chromatography column (Bio-Rad, model: Econo-Column®, catalog number: 7372576)
3. Electrophoresis system (Bio-Rad, model: Mini-PROTEAN® Tetra Cell, catalog number: 165-8004)
4. Speed-vac (Eppendorf, model: Concentrator plus, catalog number: 5305000568)
Procedure
文章信息
稿件历史记录
提交日期: Jun 6, 2025
接收日期: Jul 28, 2025
在线发布日期: Aug 11, 2025
出版日期: Sep 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Sanhueza, D. and Saez-Aguayo, S. (2025). New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage. Bio-protocol 15(17): e5433. DOI: 10.21769/BioProtoc.5433.
分类
植物科学 > 植物生物化学 > 糖类
生物化学 > 糖类 > 多糖
植物科学 > 植物分子生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












