- EN - English
- CN - 中文
Real-Time Imaging of Specific Genomic Loci With CRISPR/dCas9 in Human Cells Using CRISPRainbow
利用CRISPRainbow在人体细胞中实时成像特定位点基因组
发布: 2025年09月05日第15卷第17期 DOI: 10.21769/BioProtoc.5432 浏览次数: 3631
评审: David PaulAnonymous reviewer(s)
Abstract
Proper genome organization is essential for genome function and stability. Disruptions to this organization can lead to detrimental effects and the transformation of cells into diseased states. Individual chromosomes and their subregions can move or rearrange during transcriptional activation, in response to DNA damage, and during terminal differentiation. Techniques such as fluorescence in situ hybridization (FISH) and chromosome conformation capture (e.g., 3C and Hi-C) have provided valuable insights into genome architecture. However, these techniques require cell fixation, limiting studies of the temporal evolution of chromatin organization in detail. Our understanding of the heterogeneity and dynamics of chromatin organization at the single-cell level is still emerging. To address this, clustered regularly interspaced short palindromic repeats (CRISPR)/dead Cas9 (dCas9) systems have been repurposed for precise live-cell imaging of genome dynamics. This protocol uses a system called CRISPRainbow, a powerful tool that allows simultaneous targeting of up to seven genomic loci and tracks their locations over time using spectrally distinct fluorescent markers to study real-time chromatin organization. Multiple single-guide RNA (sgRNA), carrying specific RNA aptamers for labeling, can be cloned into a single vector to improve transfection efficiency in human cells. The precise targeting of CRISPRainbow offers distinct advantages over previous techniques while also complementing them by validating findings in live cells.
Key features
• Simultaneous imaging of up to seven specific genomic loci in living cells.
• Multicolor imaging using a single CRISPR system from Streptococcus pyogenes.
• Signal amplification through targeting repetitive sequences.
• Targeting endogenous DNA without the need for foreign DNA insertion.
Keywords: CRISPRainbowGraphical overview
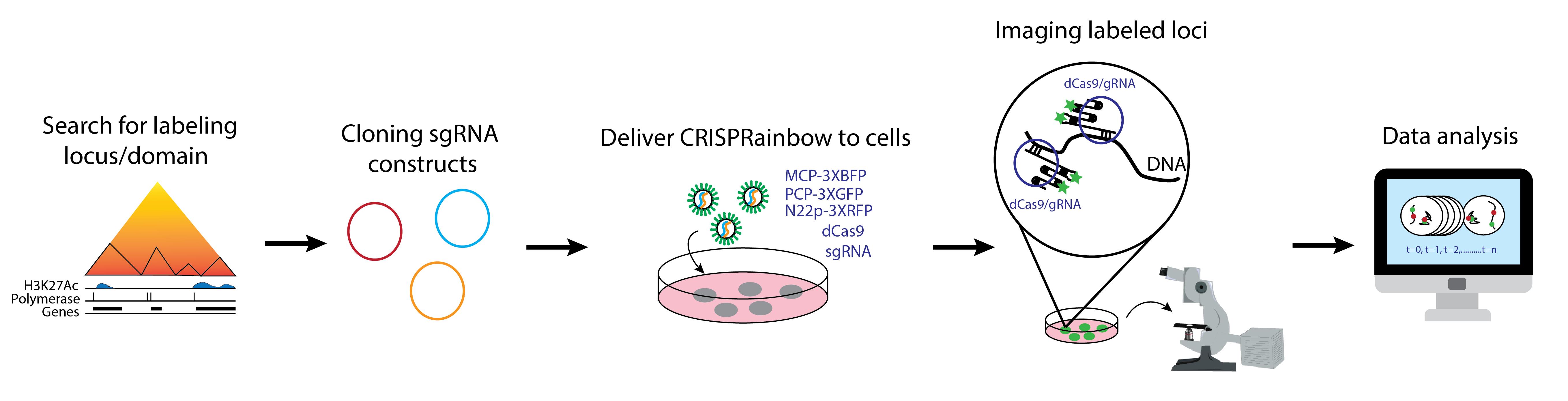
Background
CRISPR/Cas systems from Streptococcus pyogenes have been widely utilized in gene editing and silencing across various organisms owing to their ability to precisely target specific DNA sequences [1,2,3]. A particular adaptation has been the use of catalytically inactive Cas9 (dCas9), which binds DNA without introducing double-strand breaks. This enabled the repurposing of CRISPR/dCas9 into a powerful imaging platform for visualizing specific genomic loci and probing dynamic chromatin architecture in living cells [4,5,6]. Prior to the development of the CRISPR/dCas9-based imaging, studies of nuclear organization relied heavily on labor-intensive tools such as fluorescent transcription activator-like effector nucleases (TALENs) or endpoint techniques like fluorescence in situ hybridization (FISH), which are limited to fixed cells and provide only static snapshots of chromatin structure [7,8]. Although live-cell imaging using fluorescently tagged histone molecules (e.g., H2B-GFP) has provided valuable insights into global chromatin dynamics, their inability to resolve individual loci or local specific transcriptional states limits their utility in fine-scale studies of genome function. In contrast, systems such as the lac operon, which require insertion of foreign DNA elements, pose risks of artificial perturbation to the native chromatin environment. CRISPR-based imaging methods overcome many of these limitations. They allow for endogenous locus visualization without genomic modification, require minimal prior knowledge of genomic sequence context, and enable simultaneous tracking of multiple loci at the single-cell level. The ability to image chromatin structure in three dimensions and in real time opens the door to transformative insights into genome organization, transcriptional regulation, and epigenetic control—areas of immense relevance for understanding development, disease, and genome plasticity.
Labeling of genomic loci with the CRISPR/dCas9 system was initially achieved by fusing a fluorescent protein, such as EGFP, to the dCas9, allowing only one color per CRISPR system [4,5,6]. In our protocol, we engineered single-guide RNAs (sgRNAs) to carry RNA scaffolds that recruit fluorescent protein-fused RNA coat proteins for locus-specific labeling [3,9]. This method, known as CRISPRainbow, which uses spectrally separated fluorophores, facilitates multiplexed, multicolor imaging of distinct genomic regions in single cells by a single CRISPR system [9]. Such advancements greatly improve the resolution and scalability of CRISPR imaging and offer powerful complementary data when integrated with genome-wide mapping techniques like Hi-C. Altogether, CRISPR-based imaging offers unprecedented opportunities to investigate the spatial and temporal dynamics of the genome with high precision and minimal perturbation.
Materials and reagents
Biological materials
1. NEB® Stable Competent E. coli (high efficiency) (New England Biolabs, catalog number: C3040H) or One ShotTM TOP10 Chemically Competent E. coli (Thermo Fisher Scientific, catalog number: C404010), storage: -80 °C
2. One ShotTM ccdB SurvivalTM 2 T1R Competent Cells (Thermo Fisher Scientific, catalog number: A10460), storage: -80 °C
3. Human U2OS cells (ATCC, catalog number: HTB-96), storage: liquid nitrogen tank
Reagents
A. Annealing oligonucleotides
1. Custom forward and reverse oligonucleotides with standard desalting purification (Integrated DNA Technologies or similar vendors; see Section A for details), storage: -20 °C as a 100 μM stock solution. If oligos are ordered as powder, briefly spin the tubes in a minicentrifuge and dissolve the pellet in molecular biology–grade water to prepare a 100 μM solution
2. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: room temperature (RT)
3. Tris-HCl, 1 M, pH = 8.0, molecular biology grade, ultrapure (Thermo Fisher Scientific, catalog number: J22638-AP), storage: RT
4. Sodium chloride (NaCl) (Fisher Chemical, catalog number: S271-1), storage: RT
5. Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884), storage RT
B. Cloning annealed oligos into vectors
1. CRISPR sgRNA vectors with stem loops: pLH-sgRNA1-2XMS2 (Addgene, ID: 75389), pLH-sgRNA1-2XPP7 (Addgene, ID: 75390), pLH-sgRNA1-2XboxB (Addgene, ID: 75391), pLH-sgRNA1-MS2-PP7 (Addgene, ID: 75392), pLH-sgRNA1-PP7-boxB (Addgene, ID: 75393), pLH-sgRNA1-boxB-MS2 (Addgene, ID: 75396), and pLH-sgRNA1-boxB-MS2-PP7 (Addgene, ID: 75397), storage: -20 °C
2. ATP, 100 mM (New England Biolabs, catalog number: P0756S), storage: -20 °C
3. BbsI-HF® (New England Biolabs, catalog number: R3539L), storage: -20 °C
4. T7 DNA Ligase (New England Biolabs, catalog number: M0318L), storage: -20 °C
5. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
6. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
7. Zyppy® Plasmid Miniprep kit (Zymo Research, catalog number: D4019), storage: RT (except for the neutralization buffer, storage: 4 °C)
C. LB-carbenicillin plates
1. LB broth with agar (Lennox) (Sigma-Aldrich, catalog number: L2897), storage: RT
2. Carbenicillin solution, 100 mg/mL (Teknova, catalog number: C2199), storage: -20 °C
D. LB-carbenicillin liquid media
1. LB broth (Lennox) (Sigma-Aldrich, catalog number: L7658), storage: RT
E. Restriction digest of individual sgRNA
1. NotI-HF®, 20,000 U/mL (New England Biolabs, catalog number: R3189S), storage: -20 °C
2. EcoRI-HF®, 20,000 U/mL (New England Biolabs, catalog number: R3101S), storage: -20 °C
3. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
4. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
F. 1% agarose gel
1. TAE buffer, tris-acetate-EDTA, 50× (Fisher BioreagentsTM, catalog number: BP13321), storage: RT
2. Agarose (Sigma-Aldrich, catalog number: A9539), storage: RT
3. Ethidium bromide solution, 10 mg/mL (Sigma-Aldrich, catalog number: E1510), storage: RT
4. Gel loading dye, purple 6× (New England Biolabs, catalog number: B7024S), storage: -20 °C
5. 1 kb Plus DNA ladder, 500 μg/mL (New England Biolabs, catalog number: N3232L), storage: -20 °C
6. Monarch® PCR & DNA Cleanup kit (5 μg) (New England Biolabs, catalog number: T1030L), storage: RT
G. PCR amplification of individual CRISPRainbow sgRNA inserts
1. PCR primers (Integrated DNA Technologies or other vendors that synthesize single-stranded DNA oligos at 25 nmol with standard desalting purification). Dissolve the primers using molecular biology–grade water into a 100 μM stock solution, storage: -20 °C
a. For template pLH-sgRNA1-2XMS2, the forward and reverse primers are:
Forward primer sgRNA-Ex-F1: AATAATggtctcaTGACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R1: AATAATggtctcATTGCTGCCATTTGTCTCGAGGTCGAGA
b. For template pLH-sgRNA1-2XPP7, the forward and reverse primers are:
Forward primer sgRNA-In-F2: AATAATggtctcaGCAAccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R2: AATAATggtctcAGTTCTGCCATTTGTCTCGAGGTCGAGA
c. For template pLH-sgRNA1-2XboxB, the forward and reverse primers are:
Forward primer sgRNA-In-F3: AATAATggtctcaGAACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R3: AATAATggtctcAACTGTGCCATTTGTCTCGAGGTCGAGA
d. For template pLH-sgRNA1-MS2-PP7, the forward and reverse primers are:
Forward primer sgRNA-In-F4: AATAATggtctcaCAGTccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R4: AATAATggtctcAGTGTTGCCATTTGTCTCGAGGTCGAGA
e. For template pLH-sgRNA1-PP7-boxB, the forward and reverse primers are:
Forward primer sgRNA-In-F5: AATAATggtctcaACACccttcaccgagggcctatttcc
Reverse primer sgRNA-In-R5: AATAATggtctcAGACATGCCATTTGTCTCGAGGTCGAGA
f. For template pLH-sgRNA1-boxB-MS2, the forward and reverse primers are:
Forward primer sgRNA-In-F6: AATAATggtctcaTGTCccttcaccgagggcctatttcc
Reverse primer sgRNA-Ex-R1: AATAATggtctcAGAGTTGCCATTTGTCTCGAGGTCGAGA
2. dNTP, 10 mM (New England Biolabs, catalog number: N0447S), storage: -20 °C
3. Q5® Reaction Buffer pack, 5× (New England Biolabs, catalog number: B9027S), storage: -20 °C
4. Q5® high-fidelity DNA polymerase, 2,000 U/mL (New England Biolabs, catalog number: M0491L), storage: -20 °C
5. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
H. Assembling CRISPRainbow for the targets of interest
1. pCRISPRainbow-DONOR1 (Addgene, ID: 75398), storage: -20 °C
2. BsaI-HF® v2, 20,000 U/mL (New England Biolabs, catalog number: R3733L), storage: -20 °C
3. T7 DNA ligase, 3,000,000 U/mL (New England Biolabs, catalog number: M0318L), storage: -20 °C
4. ATP, 100 mM (New England Biolabs, catalog number: P0756S), storage: -20 °C
5. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
6. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
I. Restriction digest of assembled CRISPRainbow
1. ApaLI, 10,000 U/mL (New England Biolabs, catalog number: R0507S), storage: -20 °C
2. CutSmartTM buffer, 10× (New England Biolabs, catalog number: B6004S), storage: -20 °C
3. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
J. Assembled CRISPRainbow amplification
1. ZymoPURETM II Plasmid Midiprep kit (Zymo Research, catalog number D4200), storage: RT (except for ZymoPURETM P1, storage: 4 °C)
2. Corning® molecular biology–grade water (Corning, catalog number: 46-000-Cl), storage: RT
K. U2OS cell growth
1. Dulbecco’s modified Eagle medium (DMEM), 1× (Corning, catalog number: 10-013-CV), storage: 4 °C
2. Fetal bovine serum (FBS) (Gibco, catalog number: A5256701), storage: -20 °C
3. Penicillin/streptomycin (Pen/Strep, 100×), 10,000 U/mL (Gibco, catalog number: 15140122), storage: -20 °C
4. Dulbecco’s phosphate buffered saline (DPBS), 1× (Corning, catalog number: 21-030-CV), storage: 4 °C
5. TrypLETM Express enzyme, no phenol red, 1× (Gibco, catalog number: 12604013), storage: RT
L. Transfection
1. Opti-MEMTM I reduced serum medium, 1× (Gibco, catalog number: 31985062), storage: 4 °C
2. Lipofectamine® 2000 (Life Technologies, catalog number: 11668027), storage: 4 °C
3. Trypan Blue solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061), storage: RT
4. pHAGE-TO-dCas9 (Addgene, ID: 75381), storage: -20 °C
5. pHAGE-EFS-MCP-3XBFPnls (Addgene, ID: 75384), pHAGE-EFS-PCP-3XGFPnls (Addgene, ID: 75385), pHAGE-EFS-N22p-3XRFPnls (Addgene, ID: 75387), storage: -20 °C
M. Imaging
1. Olympus Type F immersion oil (Thorlabs, catalog number: MOIL-30), storage: RT
2. TetraSpeckTM microspheres, 0.1 μm, fluorescent blue/green/orange/dark red (Life Technologies, catalog number: T7279), storage: 4 °C
Solutions
1. Annealing buffer (see Recipes)
2. 1× TAE buffer (see Recipes)
3. Medium for U2OS cells (see Recipes)
Recipes
1. Annealing buffer (25 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M Tris-HCl, pH = 8.0 | 100 mM (v/v) | 0.25 mL |
| 5 M NaCl | 50 mM (v/v) | 0.25 mL |
| 0.5 M EDTA, pH = 8.0 | 1 mM (v/v) | 0.05 mL |
| ddH2O | - | 24.5 mL |
2. 1× TAE buffer (1L)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 50× TAE buffer | 2% (v/v) | 20.0 mL |
| ddH2O | 98% (v/v) | 980.0 mL |
3. Medium for U2OS cells (500 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DMEM, 1× | 89.1% (v/v) | 450 mL |
| FBS | 9.9% (v/v) | 50 mL |
| Pen/Strep, 100× | 1.0% (v/v) | 5 mL |
Laboratory supplies
1. TIPONE® pipette tips in racks, 10 μL (USA Scientific, catalog number: 1111-3200)
2. TIPONE® pipette tips in racks, 200 μL (USA Scientific, catalog number: 1110-1200)
3. TIPONE® pipette tips in racks, 1,000 μL (USA Scientific, catalog number: 1111-2721)
4. Corning® 100 mm × 20 mm style dish (Corning, catalog number: 430293)
5. 0.2 mL 8-strip PCR tube with flat cap, clear (GeneMate, catalog number: 490003-710)
6. DNA LoBind® tubes, 1.5 mL (Eppendorf, catalog number: 022431021)
7. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 5 mL (Thermo Fisher Scientific, catalog number: 13-678-11D)
8. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 10 mL (Thermo Fisher Scientific, catalog number: 13-678-11E)
9. FisherbrandTM sterile polystyrene disposable serological pipettes with magnifier stripe, 50 mL (Thermo Fisher Scientific, catalog number: 13-678-11F)
10. VWR® centrifuge tube, 15 mL (Avantor, catalog number: 525-1068)
11. VWR® centrifuge tube, 50 mL (Avantor, catalog number: 525-1075)
12. Falcon® 25 cm2 rectangular canted neck cell culture flask with plug seal screw cap (Corning, catalog number: 353082)
13. 35 mm glass bottom dishes, No. 1.5 coverslips (MatTek, catalog number: P35G-1.5-10-C)
14. L-shaped cell spreader (Thermo Fisher Scientific, catalog number: 14665230)
15. Corning Falcon round-bottom polypropylene culture tubes (Thermo Fisher Scientific, catalog number: 352059)
Equipment
1. Eppendorf Research® plus, 6-pack with Pipette Carousel 2 (Eppendorf, model: 3123000942)
2. Easypet 3 Motorized Pipette Controller (Eppendorf, model: 4430000018)
3. Mastercycler® nexus GX2 (Eppendorf, model: 6336000023)
4. EnduroTM UV Transilluminator (Labnet Internation, Inc., model: U1002-230V)
5. EnduroTM Gel XL Electrophoresis System (Labnet International, Inc., model: E0160)
6. NewClassic MF Analytical Balance (Mettler Toledo, model: MS303S)
7. Synergy® UV with integrated remote dispenser (Merck, model: 171-0774)
8. MaxQTM 6000 Incubated/Refrigerated Stackable Shakers (Thermo Fisher Scientific, model: SHKE6000-7)
9. NanoDropTM 2000c Spectrophotometer (Thermo Fisher Scientific, model: ND-2000C)
10. myECL Imager (Thermo Fisher Scientific, model: 62236X)
11. myFUGETM 12 Mini Centrifuge 100-240V (Benchmark, model: C1012)
12. Mini Vortexer (Thermo Fisher Scientific, model: 02215365)
13. CO2 Incubator (Sanyo, model: MCO-18AIC)
14. Hermle Refrigerated Centrifuge (Labnet, model: Z 400 K)
15. Class II A2 Biological Safety Cabinet (Thermo Forma, model: 1284)
16. Centrifuge (Eppendorf, model: 5424 R)
17. Large centrifuge (Eppendorf, model: 5810 R)
18. QIAvac 24 Plus (Qiagen, model: 19413)
19. Phase Contrast Microscope (Nikon, model: Diaphot 200/300, for cell culture)
20. 4 °C refrigerator
21. -20 °C refrigerator
22. -80 °C freezer
23. LocatorTM Plus Rack and Box Systems (Thermo Fisher Scientific, model: Locator 8 Plus)
24. Countess® II FL Automated Cell Counter (Invitrogen, discontinued) or Countess® 3 FL Automated Cell Counter (Invitrogen, model: A49866)
25. Epifluorescence microscope (Leica, model: DMIRB, or similar) equipped with an EMCCD camera (Andor model: iXon-897D, or similar) and 100× apochromatic objective lens (oil immersion, Olympus, model: UPLXAPO100XO, NA 1.4 or higher)
Software and datasets
1. A plasmid Editor (ApE) (M. Wayne Davis, v3.1.7) (https://jorgensen.biology.utah.edu/wayned/ape/), free to use
2. Fiji (NIH, version 2.14.0 or newer) (https://imagej.net/software/fiji/downloads), free to use
3. MetaMorph (Molecular Devices) (https://www.moleculardevices.com/products/cellular-imaging-systems/high-content-analysis/metamorph-microscopy), requires a license (Leica Microsystems)
4. Camera registration codes are available upon request to Dr. David Grunwald at the University of Massachusetts Chan Medical School.
5. OriginLab Pro (OriginLab) (https://www.originlab.com/), requires a license
Procedure
文章信息
稿件历史记录
提交日期: May 20, 2025
接收日期: Jul 29, 2025
在线发布日期: Aug 11, 2025
出版日期: Sep 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Versosky, T. J., Nishonov, D. U. and Tu, L. C. (2025). Real-Time Imaging of Specific Genomic Loci With CRISPR/dCas9 in Human Cells Using CRISPRainbow. Bio-protocol 15(17): e5432. DOI: 10.21769/BioProtoc.5432.
分类
生物物理学 > 显微技术 > 单分子定位显微技术
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












