- EN - English
- CN - 中文
Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
利用高亲和力泛素结合结构域检测和纯化泛素化底物及其互作蛋白
(*contributed equally to this work) 发布: 2025年09月05日第15卷第17期 DOI: 10.21769/BioProtoc.5426 浏览次数: 3794
评审: Kazem NouriAnonymous reviewer(s)

相关实验方案

利用基于 Western blot 的生物素化抗体内吞检测法监测膜整合蛋白的内吞过程
Alexandra Graninger and Prasanna Satpute-Krishnan
2025年11月20日 2194 阅读
Abstract
OtUBD is a high-affinity ubiquitin-binding domain (UBD) derived from a large protein produced by the microorganism Orientia tsutsugamushi. The following protocol describes a step-by-step process for the enrichment of ubiquitinated proteins from baker's yeast and mammalian cell lysates using OtUBD. The OtUBD affinity resin can strongly enrich both mono- and poly-ubiquitinated proteins from crude lysates. The protocol further describes the use of different buffer formulations to specifically enrich for proteins covalently modified by ubiquitin with or without proteins that associate with them. Combining different OtUBD-mediated enrichment protocols with liquid chromatography–tandem mass spectrometry (LC–MS/MS) helps distinguish the pool of covalently ubiquitinated proteins (the ubiquitinome) from ubiquitin- or ubiquitinated protein-interacting proteins (the ubiquitin interactome). The OtUBD tool described in the protocol has been used successfully with downstream applications such as immunoblotting and differential proteomics. It provides researchers with a versatile and economical tool for the study of ubiquitin biology.
Key features
• The protocol offers a native workflow and a denaturing workflow for enrichment of ubiquitinated proteins with or without noncovalently associated proteins, respectively.
• Included in the protocol are different resin compositions, lysate preparation methods, elution methods, and pulldown formats to suit different experimental needs.
• The protocol has been used in various applications, including immunoblotting, proteomics, and UbiCREST (ubiquitin chain restriction), and works with all types of ubiquitin conjugates.
• The protocol was developed and tested with budding yeast and mammalian cell lysates but can be adapted to other biological samples and organisms.
Keywords: Ubiquitin (泛素)Graphical overview
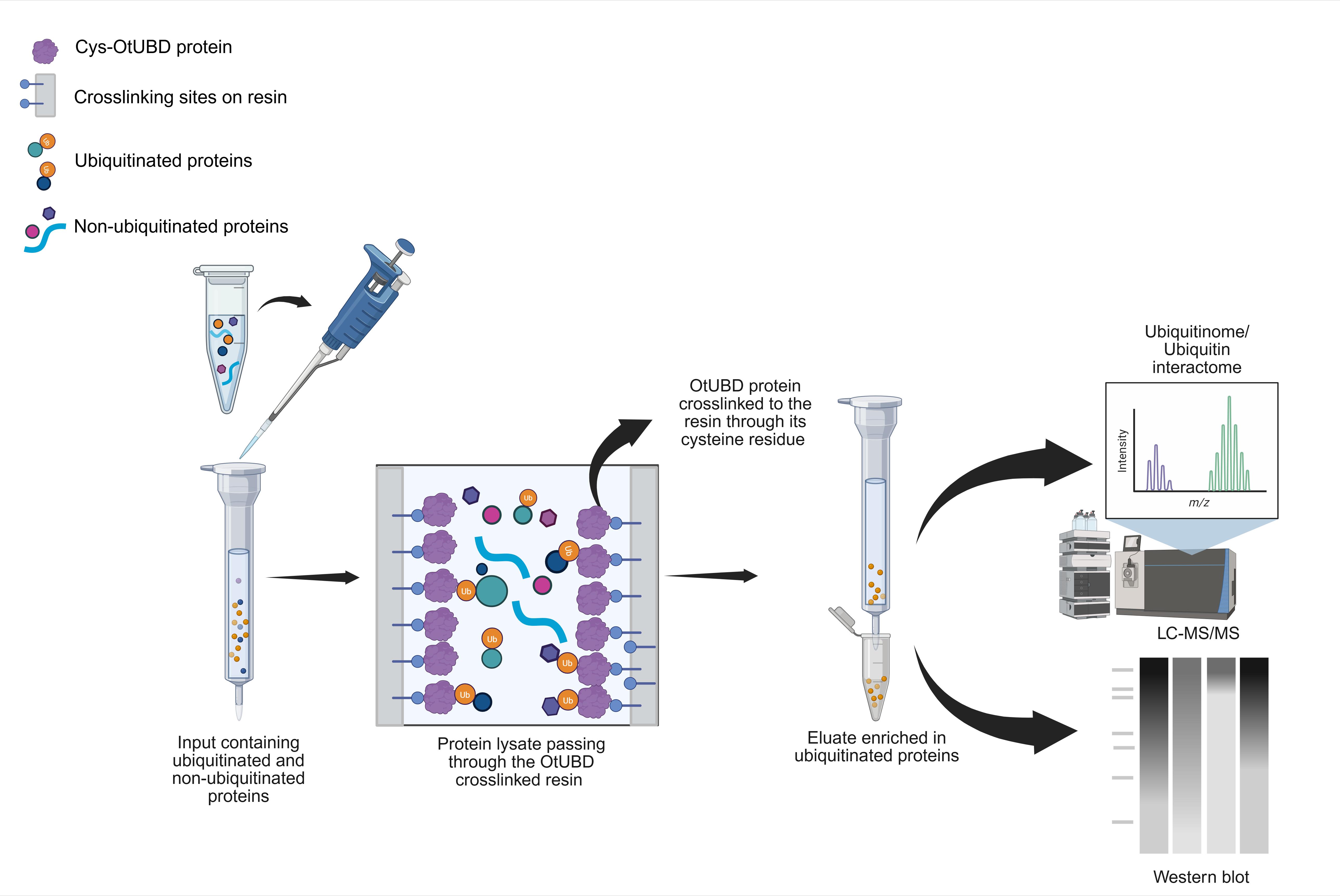
Background
The small protein ubiquitin exists in all eukaryotic organisms and is an essential post-translational protein modifier. Substrate proteins are covalently attached to ubiquitin via the coordinated reactions of three enzymes: the E1 enzyme, which mediates ATP-dependent activation of the ubiquitin C-terminus [1], the E2 conjugating enzyme, which carries the activated ubiquitin via a thioester bond [2], and finally, the E3 ligase [3], which directly interacts with the substrate and enables the transfer of ubiquitin to the substrate, which is usually, but not always, a protein [4]. Ubiquitin itself can be conjugated to other ubiquitin molecules via their N-terminal amino group or lysine side chains, forming distinct polymers that can specify interactions with different ubiquitin-binding domains [5]. For example, proteins modified by K48-linked polyubiquitin chains are typically recognized by the proteasome, where they are degraded [6], while many proteins marked with K63-linked polyubiquitin chains function in the endocytic or DNA repair pathways [7].
The diversity in ubiquitin chain topologies and linkages, as well as the attachment of single ubiquitin molecules, allows a corresponding diversity in substrate protein fates in many cellular pathways. Moreover, defective ubiquitination has been associated with many diseases, including neurodegenerative disorders and various cancers [8,9]. Thus, identifying ubiquitinated proteins and ways in which their modification might change under different conditions is often a key step in understanding cellular regulatory mechanisms and maladies resulting from their dysregulation.
At this point, many methods of enriching ubiquitinated proteins from lysates have been developed [10–12]. These each have specific advantages and disadvantages. For example, ectopic (over)expression of epitope-tagged ubiquitin, which is widely used for affinity purification of ubiquitinated targets using anti-epitope antibodies, may lead to spurious protein ubiquitination patterns, while the use of antibodies to ubiquitin for purification of conjugates at endogenous levels may lack sufficient sensitivity or specificity [13]. Tandem ubiquitin-binding entities (TUBEs), which link multiple low-affinity UBDs in a single polypeptide, are highly efficient in purifying polymeric ubiquitin, but work poorly against monoubiquitinated proteins, which usually constitute a large fraction of ubiquitinated proteins in mammalian cells and tissues [14,15]. Antibodies raised against remnant peptides derived from trypsin-digested ubiquitin conjugated to lysine side chains of substrate proteins have been extremely effective in the proteomic identification of ubiquitination sites in proteins, but the antibodies are expensive and only reveal lysine modifications and not modification of other protein side-chain sites (such as serine or threonine) or non-protein substrates [4,16].
Earlier research from our group revealed that a UBD (OtUBD) found in a large deubiquitinase protein (OtDUB) from the bacterial pathogen Orientia tsutsugamushi exhibits very high affinity for ubiquitin, with a dissociation constant in the low nanomolar range [17]. This prompted the development of an OtUBD affinity resin for the enrichment of ubiquitinated proteins from complex biological samples [18]. In the present protocol, we describe step-by-step procedures for purifying the recombinant OtUBD polypeptide and for the preparation of the OtUBD resin. We detail methods to enrich ubiquitinated proteins from either baker’s yeast or human tissue culture cells, including the use of strong denaturants to distinguish directly ubiquitinated proteins from proteins interacting noncovalently with ubiquitin or ubiquitin-modified proteins.
Materials and reagents
Plasmids used to purify OtUBD for the creation of affinity resin
1. pRT498-OtUBD (Addgene, plasmid #190089)
2. pET21a-cys-His6-OtUBD (Addgene, plasmid #190091)
Reagents
1. BactoTM peptone (Gibco, catalog number: 211677)
2. BactoTM yeast extract (Gibco, catalog number: 212750)
3. BactoTM tryptone (Gibco, catalog number: 211705)
4. Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
5. Agar (Difco, catalog number: 214010)
6. D-glucose anhydrous (Fischer chemicals, catalog number: D16-10)
7. Ampicillin sodium salt (Sigma-Aldrich, catalog number: A0166)
8. Kanamycin sulfate from Streptomyces kanamyceticus (Sigma-Aldrich, catalog number: K1637)
9. Phosphate-buffered saline (DPBS), 10×, Dulbecco's formula (Thermo Scientific, catalog number: J61917.AP)
10. Dithiothreitol (DTT) (Ultrapure AmericanBio, catalog number: AB00490)
11. DNase I (Roche, catalog number: 10104159001)
12. cOmplete EDTA-free protease inhibitor cocktail (Roche, catalog number: 11873580001)
13. Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) (Sigma-Aldrich, catalog number: E5134)
14. Glycine (Fisher Bioreagents, catalog number: BP381-5)
15. Glycerol (Sigma-Aldrich, catalog number: G7893-4L)
16. Hydrochloric acid (HCl) 36.5%–38% (J.T. Baker, catalog number: 9535-33)
17. Imidazole (Sigma-Aldrich, catalog number: I202)
18. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (RPI, catalog number: 156000)
19. L-cysteine (Sigma-Aldrich, catalog number: C7352)
20. Lysozyme (RPI, catalog number: L38100)
21. N-ethylmaleimide (NEM) (Sigma-Aldrich, catalog number: E3876)
22. Ni-NTA agarose (Qiagen, catalog number: 30230)
23. Trizma base (Tris base) for Tris buffers (Sigma-Aldrich, catalog number: T6066)
24. Tween 20 (Sigma-Aldrich, catalog number: P7949)
25. Triton-X 100 (AmericanBio, catalog number: AB02025)
26. Sodium azide (Sigma-Aldrich, catalog number: S2002)
27. SulfoLinkTM coupling resin (Thermo Scientific, catalog number: 20402)
28. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 746398)
29. Sodium dodecyl sulphate (SDS) (GFS Chemicals, catalog number: 2288)
30. Phenylmethylsulfonyl fluoride (PMSF) (MP Biomedicals, catalog number: 195381)
31. Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) (Sigma-Aldrich, catalog number: C4706)
32. Glass beads, acid washed (Sigma-Aldrich, catalog number: G8772)
33. Sodium hydroxide (NaOH) (Macron, catalog number: 7708-06)
34. GelCode Blue (Thermo Scientific, catalog number: 24594)
35. Urea (Sigma-Aldrich, catalog number: U5378)
36. PierceTM Bradford Protein Assay kit (Thermo Scientific, catalog number: 23200)
37. PierceTM BCA Protein Assay kit (Thermo Scientific, catalog number: 23225)
38. Nonfat milk (Chem Cruz, catalog number: SC-2325)
39. Immobilon-P PVDF membrane (Millipore, catalog number: IPVH00010)
40. Primary antibodies used in this study: rabbit polyclonal anti-ubiquitin antibody (Dako, Denmark, discontinued, 1:2,000 dilution), monoclonal mouse anti-ubiquitin antibody P4D1 (Enzo, USA, 1:1,000 or Invitrogen, USA, 1:4,000), and rabbit anti-ubiquitin antibody (E412J) (Cell Signalling, USA, 1:4,000)
41. For detection of rabbit primary antibodies, the HRP-linked anti-rabbit IgG secondary antibody (GE Healthcare, catalog number: NA934) was used at a dilution of 1:5,000 or 1:10,000
42. For detection of mouse primary antibodies, the HRP-linked anti-mouse secondary antibody (GE Healthcare, catalog number: NXA931V) was used at a dilution of 1:10,000
43. His-tagged tobacco etch virus (TEV) protease (purified in-house as a 2 mg/mL stock or NEB, catalog number: P8112)
44. SYPRO Ruby protein gel stain (Invitrogen, catalog number: S12000)
45. 4%–15% Mini-PROTEAN® TGXTM precast protein gels (Bio-Rad, catalog number: 4561084)
Solutions
Buffers for protein purification
1. Luria-Bertani (LB) medium (see Recipes)
2. Yeast-peptone-dextrose (YPD) medium (see Recipes)
3. 1 M TCEP (see Recipes)
4. His column buffer (see Recipes)
5. His wash buffer (see Recipes)
6. His elution buffer (see Recipes)
7. FPLC buffer (see Recipes)
8. TEV cleavage buffer (see Recipes)
Buffers for resin preparation
9. SulfoLink coupling buffer (see Recipes)
10. 50 mM L-cysteine (see Recipes)
Buffers for OtUBD resin-based purifications
11. OtUBD column buffer (see Recipes)
12. OtUBD wash buffer-1 (see Recipes)
13. OtUBD wash buffer-2 (see Recipes)
14. 1× SDS gel sample buffer (see Recipes)
15. OtUBD native lysis buffer (see Recipes)
16. OtUBD urea lysis buffer (see Recipes)
17. OtUBD elution buffer (see Recipes)
18. Neutralization buffer (see Recipes)
19. OtUBD urea wash buffer (see Recipes)
Recipes
Buffers for protein purification
1. Luria-Bertani (LB) medium
Add 5 g of yeast extract, 10 g of tryptone, and 5 g of NaCl to 1 L of MilliQ water.
Autoclave and store at room temperature.
2. Yeast-peptone-dextrose (YPD) medium
Add 10 g of yeast extract and 20 g of peptone to 950 mL of MilliQ water.
Autoclave and then add 2% glucose.
Store at room temperature.
3. 1 M TCEP
Prepare a 1 M stock solution of TCEP and neutralize it to pH 7 with 10 N NaOH.
Store aliquots at -20 °C.
4. His column buffer (for histidine tag purifications)
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole
5. His wash buffer
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole
6. His elution buffer
50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 250 mM imidazole
7. FPLC buffer
50 mM Tris-HCl, 150 mM NaCl pH 7.5, 1 mM TCEP
Prepare fresh. Filter sterilize and purge by applying a vacuum for 20 min.
8. TEV cleavage buffer
50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM TCEP
Add TCEP right before use.
Buffers for resin preparation
9. SulfoLink coupling buffer
50 mM Tris-HCl, 5 mM EDTA, pH 8.5
10. 50 mM L-cysteine
Dissolve L-cysteine in SulfoLink coupling buffer and adjust the pH to 8.5 using 10 N NaOH.
Prepare fresh before use.
Buffers for OtUBD resin-based purifications
11. OtUBD column buffer
50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerol, pH 7.5
12. OtUBD wash buffer-1
50 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20 pH 7.5
13. OtUBD wash buffer-2
50 mM Tris-HCl, 1 M NaCl pH 7.5
14. 1× SDS gel sample buffer
50 mM Tris-HCl pH 6.8, 2% SDS, 5% glycerol, 100 mM DTT, 0.005% bromophenol blue
Aliquot and store at -20 °C.
15. OtUBD native lysis buffer
50 mM Tris-HCl, 300 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, with freshly added 20 mM NEM, cOmplete mini EDTA-free protease inhibitor cocktail (Roche), and 1 mM PMSF, pH 7.5
16. OtUBD urea lysis buffer
50 mM Tris-HCl, 300 mM NaCl, 8 M urea, 1 mM EDTA, 0.5% Triton X-100, with freshly added 20 mM NEM, cOmplete mini EDTA-free protease inhibitor cocktail (Roche), and 1 mM PMSF, pH 7.5. Prepare fresh before use.
17. OtUBD elution buffer
100 mM glycine, pH 2.5 (pH adjusted with HCl)
18. Neutralization buffer
1 M Tris-HCl, pH 9
19. OtUBD urea wash buffer
Dissolve 2.93 g of urea powder in 10 mL of OtUBD native lysis buffer (without adding inhibitors) to make a final concentration of 4 M Urea. Prepare fresh before use.
Note: All the buffers are prepared at room temperature, filter-sterilized, and stored short term (weeks) at room temperature and long term (months) at 4 °C unless otherwise indicated. Filter-sterilization is not needed if all components of the buffer have been kept sterile. (Tris buffers may change in pH depending on the temperature. For the pH 7.5 Tris buffers we prepared at room temperature, the pH falls within the range of 7.5–7.8 at 4 °C.) All urea-containing buffers are prepared fresh before use or stored frozen at -20 °C to minimize urea decomposition. Unless otherwise indicated, all buffers are stable for at least 4 months when stored at 4 °C.
Laboratory supplies
1. Corning® 100 mm TC-treated culture dish (Corning, catalog number: 430167)
2. Corning® 50 mL centrifuge tubes (Millipore Sigma, catalog number: CLS430828-100EA)
3. Corning® 15 mL centrifuge tubes (Millipore Sigma, catalog number: CLS430791-500EA)
4. Amicon® Ultra centrifugal filter, 3 kDa MWCO (Millipore Sigma, catalog number: UFC9003)
5. Corning® 100 mm × 15 mm sterile Petri plates (Millipore Sigma, catalog number: CLS351029)
7. P1000, P200, and P10 pipettes (Pipetman and Eppendorf)
8. Thermo ScientificTM screw cap micro tubes (Thermo Scientific, catalog number: 3488)
9. Porcelain mortar (Capitol Scientific, catalog number: CoorsTek 66310)
10. Porcelain pestle (Capitol Scientific, catalog number: CoorsTek 66311)
11. Pyrex 2,000 mL, No. 4980 Conical flasks
12. 1.5 mL microcentrifuge tubes without caps (USA Scientific Inc., catalog number: NC9380465)
13. Borosil® tubes, test, reusable, plain end, 25 mL (Borosil®)
14. Econo-Pac® chromatography columns (Bio-Rad, catalog number: 7321010)
15. Poly-Prep® chromatography columns (Bio-Rad, catalog number: 7311550)
16. NalgeneTM Rapid-FlowTM sterile disposable bottle top filters (Thermo Scientific, catalog numbers: 595-4520 and 596-3320)
17. Corning® cell lifter (Millipore Sigma, catalog number: CLS3008)
18. InvitrogenTM flat gel loading tips (Fischer Scientific, catalog number: LC1002)
Equipment
1. Ultracold -80 °C freezer (Stirling, USA)
2. Refrigerated incubator shaker (New Brunswick Scientific, model: Innova 4230)
3. Incubator shaker (New Brunswick Scientific, model: Innova 4200)
4. Mini-PROTEANR II Cell (Bio-Rad, USA)
5. Mammalian cell 37 °C incubator with 5% CO2
6. Power pack (VWR, power source 250 V)
7. pH meter instrument (Mettler Toledo, model: SevenDirect SD20)
8. Cold room or 4 °C cabinet setup
9. Rocking shaker (Reliable Scientific, model: 55)
10. Superdex 75 gel filtration column, Hiload 16/600 (Cytiva, catalog number: 28-9893-33)
11. Sorvall Legend X1R centrifuge (Thermo Scientific, catalog number: 335650)
12. Tabletop ultracentrifuge (IEC Micromax, model: OptimaTM L-90K)
13. Mini Vortexer (VWR)
14. Rotoflex R2000 (Argos, China)
15. Misonix Sonicator 3000 with microtip probes
16. Drum roller (New Brunswick, model: TC-7)
17. G: Box imaging system with GeneSnap software
18. E-C Apparatus Corporation EC 105 (37 °C incubator)
19. Low-temperature incubator 815 (30 °C incubator)
20. Lyophilizer (Labconco, USA)
21. Incubator shaker series (New Brunswick Scientific, model: Innova44)
22. BioTek plate reader (Synergy MX)
23. ÄKTA Pure chromatography system GDM-49 (Cytiva, USA)
24. Mettler Toledo Balances (MS 3002S and MS 1045)
25. FastPrep homogenizer (MP Bio, CA, USA)
26. ChemiDoc imaging system (Bio-Rad, USA)
Software and datasets
1. ImageJ v2.14.0/1.54f
2. GraphPad Prism v8.0.1
Procedure
文章信息
稿件历史记录
提交日期: Jun 6, 2025
接收日期: Jul 22, 2025
在线发布日期: Aug 6, 2025
出版日期: Sep 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Saha, N., Zhang, M. and Hochstrasser, M. (2025). Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins. Bio-protocol 15(17): e5426. DOI: 10.21769/BioProtoc.5426.
分类
生物化学 > 蛋白质 > 免疫检测 > 免疫印迹法(WB )
系统生物学 > 蛋白质组学
生物化学 > 蛋白质 > 修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










