- EN - English
- CN - 中文
Evaluating Arabidopsis Primary Root Growth in Response to Osmotic Stress Using an In Vitro Osmotic Gradient Experimental System
利用体外渗透梯度实验系统评估拟南芥主根对渗透胁迫的响应生长
发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5397 浏览次数: 1888
评审: Tohir A. BozorovAnonymous reviewer(s)
Abstract
The root meristem navigates the highly variable soil environment where water availability limits water absorption, slowing or halting growth. Traditional studies use uniform high osmotic potentials, poorly representing natural conditions where roots gradually encounter increasing osmotic potentials. Uniform high osmotic potentials reduce root growth by inhibiting cell division and shortening mature cell length. This protocol describes a simple and effective in vitro system using a gradient mixer that generates a vertical gradient in an agar gel based on the principle of communicating vessels, exploiting gravity to generate a continuous mannitol concentration gradient (from 0 to 400 mM mannitol) reaching osmotic potentials of -1,2 MPa. It enables long-term Arabidopsis root growth analysis under progressive water deficit, improving phenotyping and molecular studies in soil-like conditions.
Key features
• Novel approach: Unique method to evaluate primary root growth in Arabidopsis under increasing osmotic potentials.
• Osmotic gradient system: Simulating a gradual osmotic gradient in the root growth zone while maintaining aerial tissues under control conditions.
• Sustained growth: Arabidopsis Col-0 and ttl1 mutant seedlings maintain proper root growth for 25 days, even at osmotic potentials as low as -1.2 MPa.
• Enhanced growth rates: Roots grown in the osmotic gradient exhibit higher growth rates than those in homogeneous high osmotic potential conditions.
• Phenotypic observation: ttl1 seedlings grown in the osmotic gradient do not show the typical swelling phenotype observed at extreme osmotic potentials (-1.2 MPa).
Keywords: Osmotic gradient (渗透梯度)Graphical overview
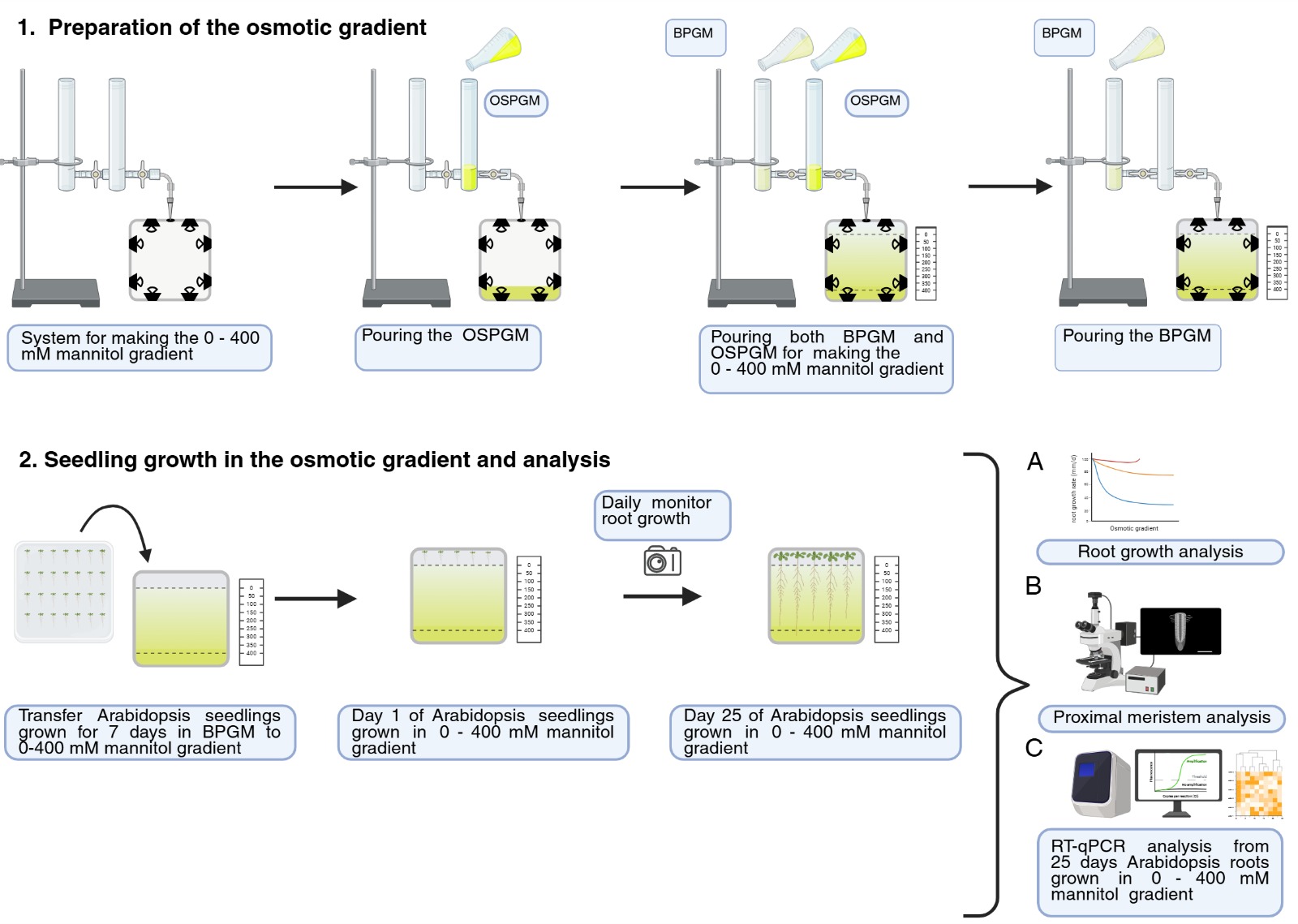
Graphical overview of the root growth analysis during osmotic stress in an in vitro osmotic gradient experimental system. BPGM: basic plant growth media; OSPMG: osmotic shock plant growth media.
Background
The root meristem faces probably one of the most complex environments on Earth: the soil, whose physicochemical properties can vary dramatically on a micrometer scale, exposing roots to various types of stress, such as osmotic stress [1]. Osmotic stress limits the ability of cells to absorb water, causing growth delay or arrest. Most studies aimed at understanding root growth adaptation to osmotic stress use homogeneous high osmotic potentials (osmotic shock) on shoots and roots [2–4]. However, this approach does not accurately mimic natural field conditions, where roots encounter progressively increasing osmotic potentials while navigating the soil. Osmotic shock significantly inhibits root growth by reducing cell division in the proximal meristem and shortening mature cell length [5]. Here, we present an efficient and straightforward protocol to generate an in vitro osmotic gradient experimental system with increasing osmotic potentials. This protocol describes a simple and effective in vitro system using a gradient mixer that generates a vertical gradient in an agar gel based on the principle of communicating vessels, exploiting gravity to generate a continuous mannitol concentration gradient (from 0 to 400 mM mannitol), reaching osmotic potentials of -1,2 MPa. The system generates a controlled osmotic gradient in the root zone while exposing the aerial tissue to control conditions. Although the use of mannitol can be discussed due to its potential toxicity, the use of high-molecular-weight polyethylene glycol, which may more closely resemble the physiological effect of drought [6], is impossible because of its incompatibility with melted agar. The gradient system has proven valuable in quantifying germination and sustained primary root growth of Arabidopsis for 25 days under continuous conditions of decreasing water availability [7]. The system could present difficulties if working for long periods because of its semi-sterile nature. The protocol paves the way for fine-tuning phenotyping and conducting molecular studies under conditions that better simulate growth in a continuum of increasing water deficit. Moreover, in the future, there will be new opportunities for studying other forms of abiotic stresses, with the modification of the described technique to create different forms of gradients.
Materials and reagents
Biological materials
1. Columbia-0 (Col-0) was used as the wild-type genotype (TAIR)
2. TTL1 (AT1G53300), T-DNA insertion line Salk (Institute Genomic Analysis Laboratory, SALK_063943 /USA)
Reagents
1. D-Mannitol (AMRESCO, catalog number: 0122)
2. Murashige and Skoog basal salt mixture (Duchefa Biocheime, catalog number: M0221.0050)
3. TRIzolTM (Invitrogen, catalog number: 15596026); store at 4 °C
4. Plant agar (Duchefa Biocheime, catalog number: P1001.1000)
5. Sucrose (Azúcar Bella Unión, catalog number: 7730106005113)
6. Kit SuperScript® IV reverse transcriptase (Invitrogen, catalog number: 18090050) using oligo d(T)20; store at -20 °C
7. DNase I (RNase-free) (NEB, catalog number: M0303S); store at -20 °C
8. PowerUpTM SYBRTM Green Master Mix (2×) (Applied Biosystems, catalog number: A25742, USA); store at 4 °C
9. EDTA disodium (Sigma-Aldrich, catalog number: 03677)
10. Ultrapure Tris (Invitrogen, catalog number: 15504-020)
11. Glacial acetic acid (Dorwill, catalog number: DA01-05-03)
12. Water for molecular biology, nuclease-free (NZYtech, catalog number: MB11101); store at 4 °C
13. RNase AWAYTM reagent (Invitrogen, catalog number: 10328011); store at room temperature
14. SyberTM Safe DNA gel stain (Invitrogen, catalog number: S33102); store at room temperature
15. TopVision agarose (Thermo, catalog number: R0492)
16. Absolute pure ethanol (Dorwil, catalog number 4403); store at -20 °C
17. TWEEN® 20 (Sigma, catalog number: P1379-100mL)
18. Sodium hypochlorite 36% (Droguería Industrial, catalog number: 10429)
19. Chloral hydrate (Sigma-Aldrich, catalog number: C8383); store in a cool, dry, well-ventilated area in a tightly closed container in an amber bottle
20. Glycerin (Sigma-Aldrich, catalog number: G5516); store in a well-sealed container at room temperature
21. Gum Arabic (Sigma-Aldrich, catalog number: G9752); store in a dry container at room temperature
22. Sterile gauze
23. Congo Red dye (Sigma-Aldrich, catalog number: C6767)
Solutions
1. Basic plant growth media (BPGM) (see Recipes)
2. Osmotic shock plant growth media (OSPGM) (see Recipes)
3. EDTA 0.5 M (see Recipes)
4. TAE 50× (see Recipes)
5. TAE 1× (see Recipes)
6. RT-qPCR reaction composition (see Recipes)
7. Agarose gel 1% (see Recipes)
8. Ethanol 70% (see Recipes)
9. Hoyer’s solution (see Recipes)
10. Sodium hypochlorite 20% (See Recipes)
Recipes
1. Basic plant growth media (BPGM) (300 mL) pH 5.7
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Murashige-Skoog basal salts (4.3 g/L) | 1× | 1.29 g |
| Plant agar | 1.2% | 3.6 g |
| Sucrose | 1.5% | 4.5 g |
| Distilled water | n/a | Up to 300 mL |
2. Osmotic shock plant growth media (OSPGM) (300 mL) pH 5.7
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Murashige-Skoog basal salts (4.3 g/L) | 1× | 1.29 g |
| Plant agar | 1.2% | 3.6 g |
| Sucrose | 1.5% | 4.5 g |
| Mannitol | 400 mM | 21.86 g |
| Distilled water | n/a | Up to 300 mL |
3. EDTA 0.5 M (250 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EDTA | 0.5 M | 46.53 g |
| Distilled water | n/a | Up to 250 mL |
Adjust to pH 8 with NaOH.
4. TAE 50× (1 L)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ultrapure Tris | 2 M | 242 g |
| Glacial acetic acid | 1 M | 57.1 mL |
| EDTA 0.5 M (pH 8) | 0.05 M | 100 mL |
| Distilled water | n/a | Up to 1,000 mL |
5. TAE 1× (1 L)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| TAE 50× | 1× | 20 mL |
| Distilled water | n/a | 980 mL |
6. RT-qPCR reaction composition (1 reaction of 10 μL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PowerUpTM SYBRTM Green Master Mix (2×) | 1× | 5 μL |
| 10 μM forward primer | 0.6 μM | 0.6 μL |
| 10 μM reverse primer | 0.6 μM | 0.6 μL |
| cDNA | ≤500 ng | 1 μL |
| Nuclease-free water | n/a | 2.8 μL |
| Total | n/a | 10 μL |
7. Agarose gel 1% (100 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Top Vision Agarose | 1% | 1 g |
| 1× TAE | n/a | 100 mL |
| SYBRTM Safe (10,000×) | 0.4× | 4 μL |
8. Ethanol 70% (1L)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Absolute pure ethanol | 70% | 700 mL |
| Distilled water | n/a | 300 mL |
9. Hoyer’s solution (100 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Chloral hydrate | 200% | 200 g |
| Gum Arabic | 30% w/v | 30 g |
| Glycerin | 20% | 20 mL |
| Distilled water | n/a | Up to 100 mL |
Store the prepared mix in a tightly sealed amber glass bottle to prevent light exposure at room temperature (around 20–25 °C) for several months.
10. 20% sodium hypochlorite
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sodium hypochlorite (36%) | 7.2% | 10 mL |
| Distilled water | n/a | Up to 50 mL |
| TWEEN 20 | n/a | 10 μL |
Laboratory supplies
1. Gradient mixer: bodies with Pyrex tubes of 20 mm external diameter, resulting in a capacity of 30 mL per tube, with 100% Teflon faucets (Uruglass Ltda., Montevideo, Uruguay) (Figure 1A)
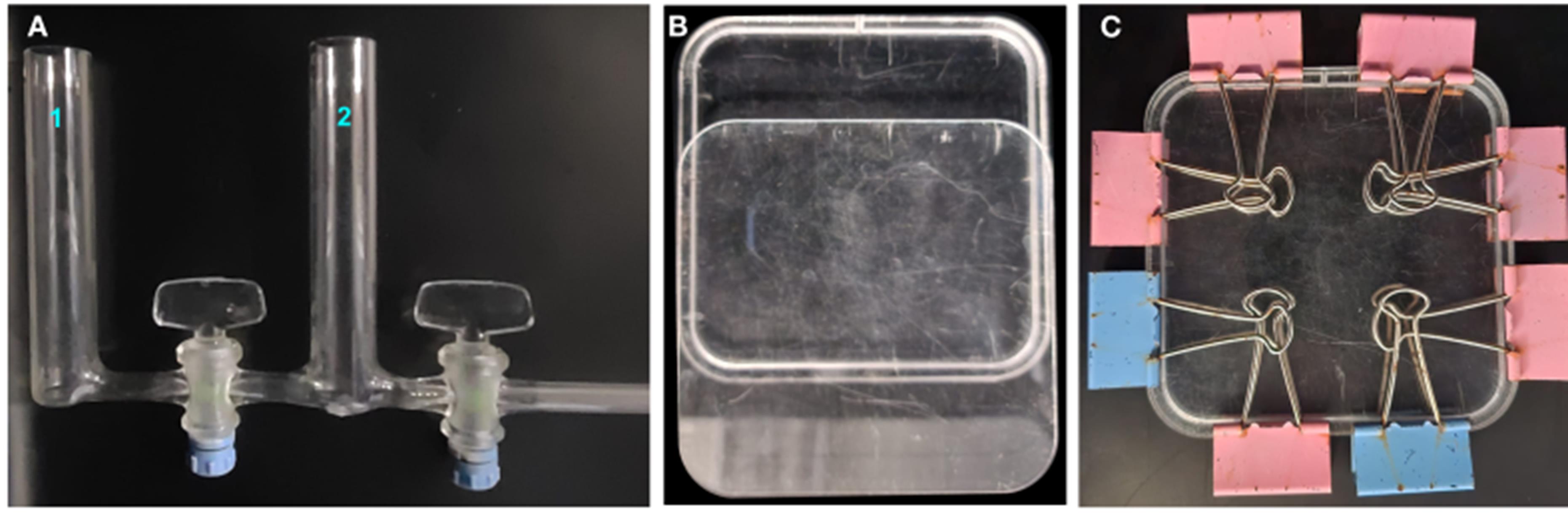
Figure 1. System components to create the osmotic gradient. (A) Gradient mixer. (B, C) Square acrylic vertical container.
2. Square acrylic vertical container with the exact dimensions of a Petri dish (120 mm width × 120 mm length × 5.7 mm depth, hole 2 mm diameter) (Figure 1B, C)
3. Square Petri dishes (120 × 120 mm) (Deltalab, catalog number: 200204)
4. 10 μL pipette tips (Tarsons, catalog number: 528100)
5. 20 μL pipette tips (Tarsons, catalog number: 528101)
6. 200 μL pipette tips (Tarsons, catalog number: 528104)
7. 1,000 μL pipette tips (Tarsons, catalog number: 528106)
8. MicroAmpTM Fast 8-tube strip, 0.1 mL (Applied Biosystems, catalog number: 4358293)
9. MicroAmpTM Optical 8-cap strips (Applied Biosystems, catalog number: 4323032)
10. Sterile 1.5 mL Eppendorf-like BIOLOGUX tubes (Biriden, catalog number:80-1500)
11. 4-way interlock flipper (SSIbio, catalog number: 5410-29)
12. Stacking 96-well PCR WorkUp rack and lead (SSIbio, catalog number: 5240-09)
13. Borosilicate glass media bottle, 500 mL, GL-45, blue caps, Schott (Droguería Paysandú)
14. Beakers borosilicate glass, 100, 200, 500, 1,000 mL (Droguería Paysandú)
15. Graduated cylinder borosilicate glass, 100, 500, 1,000 mL (Droguería Paysandú)
16. Pellet pestles (Sigma-Aldrich, catalog number: Z359963)
17. Autoclavable spatula (10 cm width)
18. PVC film (RolloPack)
19. Microscope slides and coverslips (CITOGLAS, Palmer Uruguay)
Equipment
1. Autoclave (DAIHAN Scientific, model: Fuzzy control 47Lit. MaXterile 47)
2. Horizontal laminar flow cabinet with UV lamp (Streamline® Laboratory products, Singapore)
3. Mr. FrostyTM freezing container (Thermo Fisher Scientific, catalog number: 5100-0001)
4. Liquid nitrogen (N2) tank model MVE XC20 (MVE Biological Solutions, catalog number:16520/2026)
5. Benchtop dewar flask (Thermo Fisher Scientific, catalog number: 4150-2000)
6. Freezer (-20 °C)
7. Refrigerator (2–8 °C)
8. Pipetman 4-Pipette kit, P2, P20, P200, P1000 (Gilson, catalog number: F167360)
9. Laboratory centrifuge model ST-8/8R (Thermo Fisher Scientific, catalog number: 75007204)
10. Rotor MicroClick 30×2 fixed angle microtube rotor for Sorvall ST8/ST8R (THERMO, catalog number: 75005719)
11. QuantStudioTM 5 Real-Time PCR System, 96-well, 0.1 mL (Applied Biosystems, Thermo Fisher Scientific, catalog number: A28138)
12. OACTONTM pH700 benchtop meter and stand (OACTON, catalog number: 35419-12)
13. Epifluorescence microscope, AXIO Imager, M2 with DIC (differential interference contrast) or Nomarski optics (ZEISS)
14. Digital camera (Sony, model: Cyber-shot DSC-HX1)
15. Controlled environmental chamber (PERCIVAL SCIENTIFIC, INC., USA)
16. Multifunction vortex mixer set VM-10 (DAHIAN®, catalog number: DH.WVM00020)
17. Pop-off cup head PM210 (DAHIAN®, catalog number: DH.WVM00210)
18. High-performance mini-microcentrifuge set CF-5, Class-I medical device (NIDS), Max. 5,500 rpm (DA DAIHAN®.WCF000, catalog number: DH.WCF00025) with circular fixed-angle rotor for 6× 0.2/0.5/1.5/2.0 mL tubes (DAIHAN®, catalog number: DH.WCF00105)
19. Nanodrop LITE (Thermo Scientific, catalog number: 840281500)
Software and datasets
1. ImageJ Fiji [8], free to use
2. R software [9], free to use
3. Zeiss ZENpro-Imaging Software, requires a license
4. Design and analysis software 2.7.0 QuantStudioTM (Applied Biosystems, Thermo Fisher Scientific), requires a license
5. InfoStat versión 2011 [10]
6. Excel Microsoft Corporation (2024) [11]
7. RT-qPCR results: ttl_col_dataset.xls (File S1)
8. Root growth rate data sets from osmotic gradient: data.RGR-G.xls (File S2); osmotic shock: data.RGR-S.xls (File S3)
Code
library(tidyverse)# Read datattl0 <- read_csv(file = 'ttl_col_dataset.csv', col_names = T) %>% mutate(Sample = paste(Genotype, Condition, sep = '_'))ttl_df <- ttl0 %>% dplyr::select(c(Sample, Gene, Log2FC, Rep, Condition, Genotype)) %>% pivot_wider(names_from = 'Gene', values_from = 'Log2FC')# Distances Heatmap -------------------------------------------------------library("pheatmap")ttl_mat <- ttl_df[,-c(1,2,3,4)] %>% as.matrix()rownames(ttl_mat) <- paste(rep(c('Col-0', 'ttl1'), each = 9), rep(rep(c('Control', 'Osmotic Shock', 'Osmotic Gradient'), each = 3), 2))# Distances Matrixdist_mat <- dist(ttl_mat)sampleDistMatrix <- as.matrix(dist_mat)colors <- colorRampPalette( rev(brewer.pal(9, "Blues")) )(255)# Plotpheatmap(sampleDistMatrix, clustering_method = 'complete', col=colors, # labels_row = T, show_colnames = F)# PCA ---------------------------------------------------------------------library(FactoMineR)library(factoextra)# Tidy data for prcompttl_pca <- ttl_df[, -c(1,2,3,4)]# Dimension reduction using PCAres.pca <- prcomp(ttl_pca, scale = TRUE)## Biplotfviz_pca_biplot(res.pca, title = '', axes = c(1,2), habillage = interaction(ttl_df$Condition, ttl_df$Genotype), addEllipses = T, ellipse.type = 'confidence', col.ind = ttl_df$Condition, label = 'var', pointshape = 19, mean.point = T)+ geom_point(aes( shape = ttl_df$Genotype), size = 2)+ scale_color_manual(name = 'Conditions', labels = rep(c('Control', 'Gradient', 'Shock'), times = 2), values = c(paleta, paleta))+ scale_fill_manual(name = 'Conditions', labels = rep(c('Control', 'Gradient', 'Shock'), times = 2), values = c(paleta, paleta))+ scale_shape_discrete(name = 'Genotype', labels = c('Col-0', 'TTL1'))+ theme_bw(base_family = 'Arial')+ theme( legend.position = "bottom", legend.box = 'horizontal', legend.title = element_text(size = 11), legend.text = element_text(size = 10), axis.text.x = element_text(size = 10), axis.text.y = element_text(size = 10), axis.title.x = element_text(size = 12), axis.title.y = element_text(size = 12))# Heatmap -----------------------------------------------------------------library(RColorBrewer)library(gplots)heatmap_df <-ttl0 %>% group_by(Sample, Gene) %>% summarise(mLfc = median(Log2FC)) %>% pivot_wider(names_from = 'Gene', values_from = 'mLfc')heatmap_mat <- heatmap_df[,-1] %>% as.matrix()rownames(heatmap_mat) <- heatmap_df %>% pull(Sample)# Matrix for plotmatPlot <- t(heatmap_mat)ComplexHeatmap::pheatmap(matPlot, color = redgreen(75), # color = redblue(75), scale = 'row', cluster_cols = F, cluster_rows = clust_plot, # clustering_distance_rows = clust_plot, clustering_method = 'complete', gaps_col = 3, cutree_rows = 4, column_names_side = 'top', angle_col = '45')# Packageslibrary (emmeans)library (lme4)library(tidyverse)#Read data RGR #data.RGR.G <- read_delim (file ="data.RGR.G.csv", col_names = TRUE, delim = ",", na = "NA")data.RGR.S <- read_delim (file ="data.RGR.S.csv",col_names = TRUE, delim = ",", na = "NA"# Data analysismod.RGR.G <- lm (RGR ~ genotype + medium + genotype * medium, data = data.RGR.G)anova (mod.RGR.G)em.RGR.G <- emmeans (mod.RGR.G, ~ genotype * medium)cr.em.RGR.G <- contrast (em.RGR.G, method = "pairwise")mod.RGR.S <- lm (RGR ~ genotype + medium + genotype * medium, data = data.RGR.S)anova (mod.RGR.S)em.RGR.S <- emmeans (mod.RGR.S, ~ genotype * medium)cr.em.RGR.S <- contrast (em.RGR.S, method = "pairwise")Procedure
文章信息
稿件历史记录
提交日期: Mar 31, 2025
接收日期: Jun 24, 2025
在线发布日期: Jul 13, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Píriz-Pezzutto, S., Martínez-Moré, M., Sainz, M. M., Borsani, O. and Sotelo-Silveira, M. (2025). Evaluating Arabidopsis Primary Root Growth in Response to Osmotic Stress Using an In Vitro Osmotic Gradient Experimental System. Bio-protocol 15(14): e5397. DOI: 10.21769/BioProtoc.5397.
分类
植物科学 > 植物生理学 > 非生物胁迫
植物科学 > 植物生理学 > 植物生长
生物信息学与计算生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link