- EN - English
- CN - 中文
Flow Cytometric Quantification of HIV-1-Infected Cells Expressing Either Abortive or Elongated HIV-1 Transcripts Using Flow-FISH
基于Flow-FISH的流式细胞术检测终止型与延伸型HIV-1转录本阳性细胞的定量分析
(*contributed equally to this work) 发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5392 浏览次数: 2143
评审: Andrea GramaticaAndrew SylwesterAnonymous reviewer(s)
Abstract
The persistence of the HIV-1 reservoir remains the ultimate obstacle in achieving a cure. Cure strategies targeting the HIV-1 reservoir are under development, and therefore, finding ways to improve the detection of the reservoir is crucial. Several reservoir detection techniques exist to assess different markers of the HIV-1 reservoir, such as PCR-based assays and protein-based flow cytometric methods. We developed a flow cytometry-fluorescent in situ hybridization (flow-FISH) approach that assesses HIV-1 at the transcriptional level. Using a combination of probes that target either the HIV-1 trans-activation response (TAR) region and 5′ long terminal repeat (LTR) or the Gag sequence, our assay distinguishes between infected cells expressing abortive or elongated HIV-1 RNAs. This assay utilizes the branched-DNA method to amplify the fluorescent signal of the hybridized RNA probes and can be used directly for thawed or cultured cells, with the option to include surface antibody staining. Cellular expression of abortive and/or Gag HIV-1 RNAs is measured by flow cytometry. Our flow-FISH approach gives insight into the transcriptional dynamics of the HIV-1 reservoir and allows for the characterization of latently infected cells.
Key features
• Detection of latently HIV-1-infected cells identified by the expression of abortive HIV-1 TAR transcripts.
• Cell activation is not required for HIV-1 detection; therefore, the cellular phenotypic landscape remains preserved.
• Can be used for direct ex vivo measurements in isolated cells, such as PBMCs, from untreated and antiretroviral therapy (ART)-treated people with HIV-1 (PWH).
Keywords: HIV-1 reservoir (HIV-1潜伏库)Graphical overview
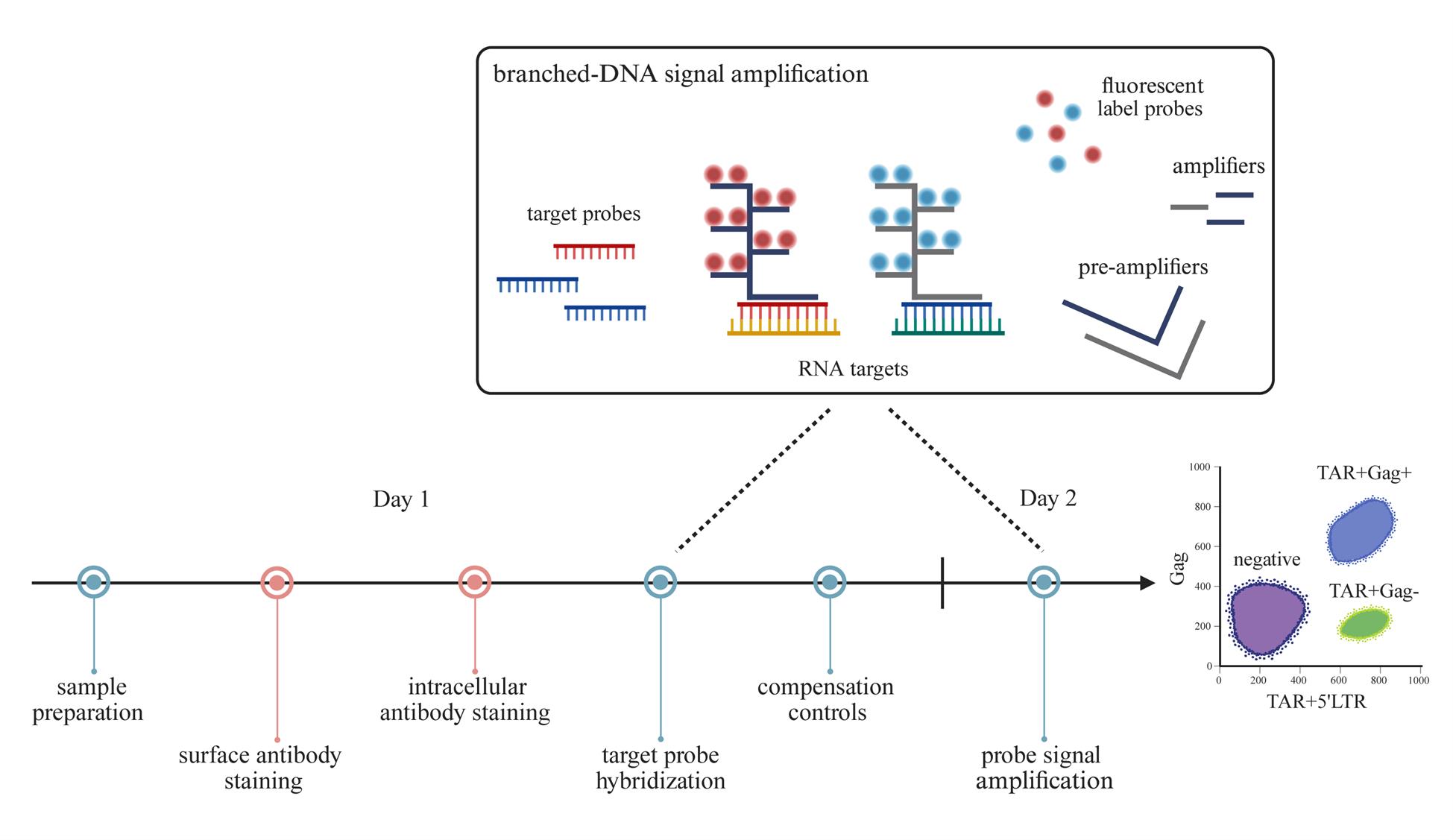
Background
Detecting and monitoring the HIV-1 reservoir of people with HIV-1 (PWH) is crucial in advancing cure strategies. Various detection methods quantify the viral reservoir based on the different characteristics of the provirus (i.e., defect, intact, transcriptionally active, translationally active, or replication-competent provirus) [1–6]. The lack of viral antigen expression associated with HIV-1 latency complicates the assessment of the viral reservoir at the cellular level. To address this problem, we developed an assay capable of detecting these latently infected cells by targeting abortive HIV-1 RNA. Inefficient transcriptional elongation of the HIV-1 genome can lead to the production of short abortive transcripts that comprise the TAR region—a highly conserved RNA sequence found at the 5′ end of HIV-1 transcripts. These abortive TAR transcripts have been identified in latency cell lines and in resting CD4+ T cells from antiretroviral therapy (ART)-treated PWH [7–9]. We thus designed a flow cytometry–fluorescent in situ hybridization (flow-FISH) assay, in which flow cytometry is combined with RNA-binding probes and branched-DNA technology to amplify the fluorescent signal. We designed one probe that targets the trans-activation response (TAR) region to detect all HIV-1 RNAs, including abortive transcripts, and another probe that targets the Gag region for the detection of elongated HIV-1 RNAs. These probes allow for flow cytometric quantification of HIV-1-infected cells expressing only abortive TAR RNAs or elongated HIV-1 RNAs—a proxy for active HIV-1 transcription. In addition, we designed a third probe that binds the 5′ long terminal repeat (5′LTR; R-U5)—a sequence directly downstream of TAR but upstream of Gag—to boost the TAR probe signal for abortive transcripts with a block to elongation. It is important to note that these probe combinations do not differentiate between cells containing replication-competent and defective proviruses. Nevertheless, our flow-FISH assay is a useful tool in monitoring the HIV reservoir and can be used to phenotypically characterize latently infected cells by flow cytometry. While this protocol is optimized for peripheral blood mononuclear cells (PBMCs), this assay can also be adapted for the assessment of the viral reservoir within lymphoid tissues of PWH.
Materials and reagents
Biological materials
1. ACH-2, a latently HIV-1-infected T-cell line derived from CEM cells (NIH AIDS Reagent Program; RRID: CVCL_0138)
2. PBMCs from PWH and an HIV-negative blood donor; isolated fresh or frozen
Reagents
1. Target probe sets, 20× (custom-made, Affymetrix, Santa Clara, CA, USA; see Table S1 for used target sequences from HIV-1 clade B); store at -20 °C
a. HIV-1 TAR in Alexa Fluor 647
b. HIV-1 5′LTR in Alexa Fluor 647
c. HIV-1 Gag in Alexa Fluor 488
2. Primeflow RNA Assay kit (Invitrogen, catalog number: 88-18005), containing:
a. RNA tubes (1.5-mL microcentrifuge tubes) (Invitrogen, catalog number: 19197)
b. Fixation buffer 1A (Invitrogen, catalog number: 00-18100); store at 2–8 °C
c. Fixation buffer 1B (Invitrogen, catalog number: 00-18200); store at 2–8 °C
d. Fixation buffer 2 (8×) (Invitrogen, catalog number: 00-18400); store at 2–8 °C
e. Permeabilization buffer (10×) (Invitrogen, catalog number: 00-18300); store at 2–8 °C
f. RNase inhibitor (100×) (Invitrogen, catalog number: 00-16002); store at 2–8 °C
g. Wash buffer (Invitrogen, catalog number: 00-19180); store at 2–8 °C
h. Target probe diluent (Invitrogen, catalog number: 00-19185); store at 2–8 °C
i. PreAmp mix (Invitrogen, catalog number: 00-16000); store at 2–8 °C
j. Amp mix (Invitrogen, catalog number: 00-16001); store at 2–8 °C
k. Label probe diluent (Invitrogen, catalog number: 00-19183); store at 2–8 °C
l. Label probes (100×) (Invitrogen, catalog number: 00-16003); store at -20°C
m. Intracellular (IC) fixation buffer (Invitrogen, catalog number: 00-8222); store at 2–8 °C
n. Storage buffer (Invitrogen, catalog number: 00-19178); store at 2–8 °C
o. Compensation kit (Invitrogen, catalog number: 88-17009)
3. UltraComp eBeadsTM microspheres (Thermo Fisher Scientific, Invitrogen, catalog number: 01-2222); store at 2–8 °C
4. PrimeFlowTM Compensation Control Alexa FluorTM 647 (Invitrogen, catalog number: PF51-17002); store at 2–8 °C
5. PrimeFlowTM Compensation Control Alexa FluorTM 488 (Invitrogen, catalog number: PF51-17003); store at 2–8 °C
6. Primeflow microRNA pretreatment buffer (Invitrogen)
a. Pretreatment concentrate (4×) (Invitrogen, catalog number: 00-16009); store at 2–8 °C
b. Pretreatment diluent (Invitrogen, catalog number: 00-16008); store at 2–8 °C
7. CD4-PE antibody (RPA-T4 clone) (BioLegend, catalog number: 300507); store at 2–8 °C
8. TNF-α (PeproTech, catalog number: 300-01A); store at -20 °C
9. Phosphate-buffered saline (PBS), 1×, pH ~7.4 (Thermo Fisher Scientific, catalog number: 10010023); store at room temperature
10. Bovine serum albumin (BSA) (Merck Sigma-Aldrich, catalog number: A7906); store at room temperature
11. Ethylenediaminetetraacetic acid (EDTA) (Thermo Fisher Scientific, catalog number: 17892); store at room temperature
12. Sodium azide (Merck, Sigma-Aldrich, catalog number: S2002); store at room temperature
Solutions
1. FACS buffer (see Recipes)
Recipes
1. FACS buffer (store at 4 °C)
| Reagent | Final concentration |
|---|---|
| PBS | |
| BSA | 0.3% |
| EDTA | 2 mM |
| Sodium azide | 0.01% |
Equipment
1. FACSCanto II device with lasers 488 nm, 561 nm, and 633 nm (BD Biosciences, Franklin Lakes, NJ, USA)
2. Centrifuge (ROTANTA 460R, Hettich, Kirchlengern, Germany); swinging bucket with adaptors for 15 mL conical tubes; acceleration 9, deceleration 7
3. Refrigerated centrifuge for 1.5 mL Eppendorf tubes (Eppendorf, model: Centrifuge 5430 R)
4. Heat block for 1.5 mL Eppendorf tubes (QB Series Dry Block Heating Systems, Grant Instruments, Cambridge, England)
5. Vortex mixer (Reax top, Heidolph Scientific Products, Schwabach, Germany)
Software and datasets
1. FlowJo software version 10; application for flow cytometry data analysis (TreeStar, Ashland, OR, USA)
2. BD FACSDiva software; a collection of tools for flow cytometer and application setup, data acquisition, and data analysis (BD Biosciences)
Procedure
文章信息
稿件历史记录
提交日期: Apr 16, 2025
接收日期: Jun 16, 2025
在线发布日期: Jul 3, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Man, S., Geijtenbeek, T. B.H. and Kootstra, N. A. (2025). Flow Cytometric Quantification of HIV-1-Infected Cells Expressing Either Abortive or Elongated HIV-1 Transcripts Using Flow-FISH. Bio-protocol 15(14): e5392. DOI: 10.21769/BioProtoc.5392.
分类
微生物学 > 病原体检测 > FISH
细胞生物学 > 细胞染色 > 核酸
细胞生物学 > 基于细胞的分析方法 > 流式细胞术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link