- EN - English
- CN - 中文
Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR
利用RATE-PCR全面定位EZ-Tn5转座子在阿根廷假单胞菌SA190中的插入位点
发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5389 浏览次数: 1759
评审: Joyce ChiuAnonymous reviewer(s)
Abstract
Transposon mutagenesis is a powerful tool for investigating gene function in bacteria, particularly in newly discovered species. In this study, we applied the hyperactive EZ-Tn5 transposase system to Pseudomonas argentinensis SA190, an endophytic bacterium known for enhancing plant resilience under drought stress. By leveraging the random amplification of transposon ends (RATE)-PCR method, we successfully mapped the insertion sites of the transposon within the SA190 genome. This approach enabled the precise identification of disrupted genes, offering insights into their roles in bacterial function and interaction with host plants. Our comprehensive protocol, including competent cell preparation, transformation, and insertion site mapping, provides a reliable framework for future studies aiming to explore gene function through mutagenesis.
Key features
• The use of the hyperactive EZ-Tn5 transposase system ensures efficient and detectable random mutagenesis across the Pseudomonas argentinensis SA190 genome, facilitating comprehensive gene disruption studies.
• The technique is employed to identify and map the transposon insertion sites, allowing for precise determination of gene function and its impact on bacterial phenotypes.
• This method enables the exploration of a broad range of gene functions within SA190, particularly those involved in plant growth promotion and stress tolerance.
• This method can be readily adapted to generate mutant libraries in other bacterial species, emphasizing its transferability.
Keywords: Transposon mutagenesis (转座子诱变)Graphical overview
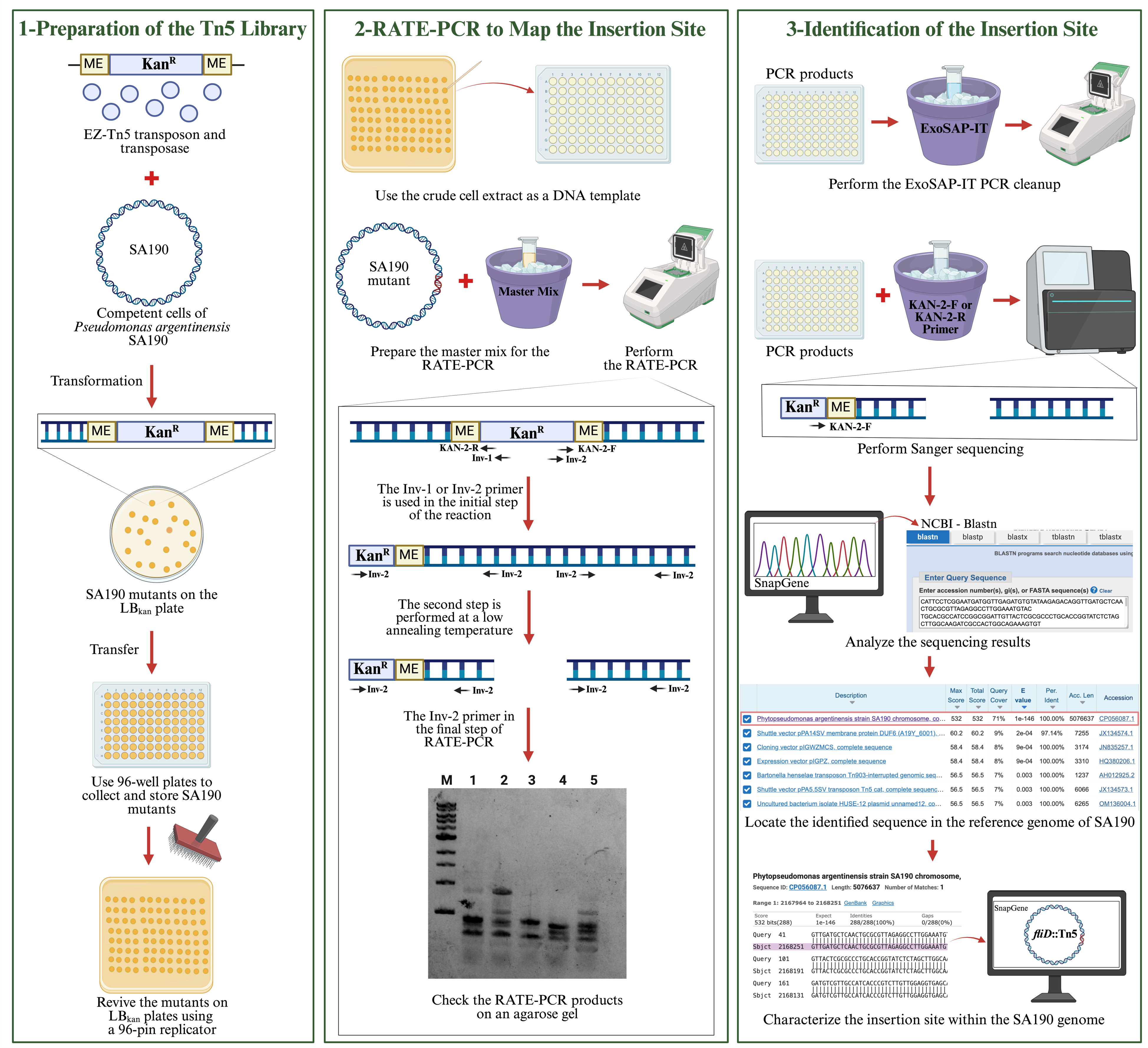
Workflow for characterizing the Tn5 insertion site in the Pseudomonas argentinensis SA190 genome using RATE-PCR
Background
Identification of essential genes by bacterial genomic studies helps to understand the functions of biological processes in bacteria [1–4]. One of the indispensable tools for studying genes in bacteria is mutagenesis, e.g., transposons insertion [1,5]. It allows us to establish a link between gene disruption and phenotypic traits. Tn5 is one of the most common transposases used to generate random mutations in the bacterial genome [6]. However, random mutagenesis methods vary in precision and efficiency, with chemical and UV mutagenesis often causing multiple mutations [7], while conventional transposons frequently exhibit lower insertion efficiency and can result in multiple insertions per genome [8]. The EZ-Tn5 transposome system addresses these limitations by incorporating a hyperactive Tn5 transposase and a kanamycin resistance cassette, allowing high-frequency, single-copy insertions that simplify mutant analysis [4,8]. Its broad host range and proven reliability across diverse bacterial species make EZ-Tn5 the preferred tool for generating effective mutant libraries in this study [8].
Mechanistically, the EZ-Tn5 transposome system, developed from the native Tn5 structure, consists of hyperactive Tn5 transposase and a kanamycin antibiotic cassette [6,9]. In the EZ-Tn5 system, the hyperactive transposase binds to the mosaic ends at the ends of the kanamycin cassette and facilitates random transposition in the genome [6,9]. The presence of the kanamycin cassette facilitates the selection of bacteria in which transposition occurs. After transposition takes place in the bacterial genome, the transposase decays [9]. In this way, transposition occurs only once in the genome.
In this work, the EZ-Tn5 transposon system was applied on the bacteria Pseudomonas argentinensis SA190, an endophytic plant growth-promoting bacteria that enhances plant performance under drought stress [10]. As SA190 is a newly discovered bacterium, it is crucial to understand its essential genes. Elucidating the role of genes in SA190 will contribute to understanding the beneficial interaction between SA190 and plants. SA190 should first be prepared as competent cells to receive the transposon DNA for transposon mutagenesis. The transposon is transferred to recipient-competent cells by transformation. Since EZ-Tn5 performs random mutagenesis in the bacterial genome, identifying the mutated gene is vital in establishing a significant relationship between the gene and its function. To achieve this, we employed the random amplification of transposon ends (RATE)-PCR method, a fast and effective approach for identifying disrupted genes in the genome [11,12]. Specific primers designed for the transposon ends were used to amplify one of the transposon-end regions. RATE-PCR involves three stages, utilizing a combination of high-stringency and low-stringency temperatures to produce both specific and nonspecific PCR products of various sizes on agarose gels. These products were then subjected to Sanger sequencing using either the forward or reverse primers of the EZ-Tn5 transposon. The resulting sequences were compared against the SA190 reference genome to precisely characterize the disrupted genes.
Materials and reagents
Biological materials
1. Pseudomonas argentinensis SA190 (isolated from surface-sterilized root nodules of the plant Indigofera argentea in the Jizan area (16°56.475′N, 42°36.694′E) of Saudi Arabia [13], kindly provided by Heribert Hirt Lab at King Abdullah University of Science and Technology (KAUST), Jeddah, Saudi Arabia).
Reagents
1. Kanamycin sulfate (Fisher Bioreagents, catalog number: 25389-94-0)
2. BactoTM tryptone (Gibco, catalog number: 211705)
3. Sodium chloride (NaCl) (Merck, catalog number: 1064049025)
4. BactoTM yeast extract (Gibco, catalog number: 212750)
5. Sodium hydroxide (NaOH) (Merck, catalog number: 71687-500G)
6. LB broth with agar (Sigma-Aldrich, catalog number: L2897-250G)
7. Potassium chloride (KCI) (Merck, catalog number: P9541-500G)
8. Magnesium chloride (MgCI2) (Merck, catalog number: 208337-100G)
9. Magnesium sulfate (MgSO4) (Merck, catalog number: M7506-500G)
10. Glucose (Merck, catalog number: G8270-100G)
11. Absolute glycerol (Sigma-Aldrich, catalog number: G5516)
12. Nuclease-free water (New England Biolabs, catalog number: B1500S)
13. Phusion® High-Fidelity DNA polymerase (New England Biolabs, catalog number: M0530L)
14. ExoSAP-IT PCR product cleanup reagent (Thermo Fisher Scientific, catalog number: 78-201-1ML)
15. 10 mM dNTPs (Thermo Fisher Scientific, catalog number: 18427089)
Solutions
1. Luria-Bertani (LB) agar (see Recipes)
2. Luria-Bertani (LB) media (see Recipes)
3. Super optimal broth with catabolite repression (SOC) media (see Recipes)
4. Kanamycin sulfate stock solution (50 mg/mL) (see Recipes)
Recipes
1. Luria-Bertani (LB) agar
| Reagent | Final concentration | Quantity |
|---|---|---|
| Tryptone | 1% (w/v) | 10 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| NaCl | 1% (w/v) | 10 g |
| Agar | 0.9% | 9 g |
| Milli-Q H2O | variable | Fill to 1 L |
| Total | n/a | 1 L |
a. Adjust pH to 7.0 using 5 M NaOH
b. Autoclave (121 °C, 15 min, 100.8 kPa).
2. Luria-Bertani (LB) media
| Reagent | Final concentration | Quantity |
|---|---|---|
| Tryptone | 1% (w/v) | 10 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| NaCl | 1% (w/v) | 10 g |
| Milli-Q H2O | variable | Fill to 1 L |
| Total (optional) | n/a | 1 L |
a. Adjust pH to 7.0 using 5 M NaOH
b. Autoclave (121 °C, 15 min, 100.8 kPa).
3. Super optimal broth with catabolite repression (SOC) media
| Reagent | Final concentration | Quantity |
|---|---|---|
| Tryptone | 2% (w/v) | 20 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| NaCl | 10 mM | 2 mL |
| KCl | 2.5 mM | 2.5 mL |
| MgCl2 | 10 mM | 10 mL |
| MgSO4 | 10 mM | 10 mL |
| Milli-Q H2O | variable | Fill to 980 mL |
| Total | n/a | 980 mL |
a. Autoclave (121 °C, 15 min, 100.8 kPa).
b. After autoclaving, add 20 mL of filter-sterilized 1 M glucose.
4. Kanamycin sulfate stock solution (50 mg/mL)
| Reagent | Final concentration | Quantity |
|---|---|---|
| Kanamycin sulfate | 50 mg/mL | 0.5 g |
| Milli-Q H2O | variable | 10 mL |
| Total | n/a | 10 mL |
a. Filter sterilize the solution using a 0.2 μm filter and 10 mL syringe.
b. Aliquot 500 μL stocks and store at -20 °C.
Laboratory supplies
1. 1.5 mL Eppendorf tubes (Thermo Fisher Scientific, catalog number: AM12450)
2. 2 mL Eppendorf tubes (Thermo Fisher Scientific, catalog number: AM12475)
3. NuncTM Petri dishes (Thermo Fisher Scientific, catalog number: 263991)
4. NuncTM square BioAssay dishes (Thermo Fisher Scientific, catalog number: 240845)
5. Inoculating loop (VWR®, catalog number: 612-7277)
6. L-shaped spreader (VWR®, catalog number: 612-1561)
7. 50 mL conical tubes (Thermo Fisher Scientific, catalog number: AM12502)
8. 0.1 cm electroporation cuvette (Thermo Fisher Scientific, catalog number: P41050)
9. NalgeneTM polycarbonate Erlenmeyer flask (Thermo Fisher Scientific, catalog number: 4103-0250PK)
10. Serological pipette (Thermo Fisher Scientific, catalog number: 170358T)
11. EZ-Tn5TM <KAN-2>
Note: This product is no longer available; as an alternative, we suggest using EZ-Tn5 Transposase (LGC Biosearch Technologies, catalog number: TNP92110)
12. CorningTM CostarTM 96-well plate, sterile (Fisher Scientific, catalog number: 10695951)
13. PCR tubes (Thermo Fisher Scientific, catalog number: AB0490)
14. 0.2 μm FisherbrandTM syringe filters, sterile (Thermo Fisher Scientific, catalog number: 09-719C)
15. 10 mL BD Luer-LokTM disposable syringes without needles (Thermo Fisher Scientific, catalog number: 14-823-2A)
16. Toothpick (local purchase)
Equipment
1. MicroPulser Electroporator (Bio-Rad Laboratories, catalog number: 1652100)
2. Centrifuge 5430 (Eppendorf, catalog number: 5427000610)
3. New Brunswick Innova® 40/40R Benchtop Orbital Shaker (Eppendorf, catalog number: M1299-0090)
4. HerathermTM General Protocol Microbiological Incubator (Thermo Fisher Scientific, catalog number: 51028063)
5. SimpliAmpTM Thermal Cycler (Thermo Fisher Scientific, catalog number: A24811)
6. 96-Pin Microplate Replicator (Boekel Scientific, catalog number: 140500)
7. EppendorfTM ResearchTM plus, mechanical single-channel pipettes multipacks (Thermo Fisher Scientific, catalog number: 13-684-250)
8. Autoclave (any manufacturer)
9. Thermo ScientificTM RevcoTM ExF -86 °C upright ultra-low temperature freezer (Thermo Fisher Scientific, catalog number: EXF24086A)
10. Liquid nitrogen (N2) tank (any manufacturer)
Software and datasets
1. SnapGene® (SnapGene, Version 6.2.1, https://www.snapgene.com/)
2. NCBI Blastn program, free use, web-based platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi)
Procedure
文章信息
稿件历史记录
提交日期: Apr 9, 2025
接收日期: Jun 19, 2025
在线发布日期: Jul 9, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Elkatmis, B., Han, B., Parween, S., Kopriva, S., Hirt, H. and Saad, M. M. (2025). Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR. Bio-protocol 15(14): e5389. DOI: 10.21769/BioProtoc.5389.
分类
微生物学 > 微生物遗传学 > 诱/突变
分子生物学 > DNA > 诱/突变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













