- EN - English
- CN - 中文
Metabolite Production and Extraction of Indole Compound From the Tomato Endophyte Streptomyces sp. VITGV100
番茄内生菌Streptomyces sp. VITGV100中吲哚类代谢产物的合成与提取分析
发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5386 浏览次数: 2379
评审: Samik BhattacharyaAnonymous reviewer(s)

相关实验方案

非洲地区疫苗衍生脊髓灰质炎病毒(cVDPV)暴发的演化与传播可视化分析
D. Collins Owuor [...] Anfumbom K.W. Kfutwah
2025年07月05日 1847 阅读
一种快速且经济的流程,用于从细菌草图基因组中识别并捕获生物合成基因簇
Marco A. Campos-Magaña [...] Luis Garcia-Morales
2025年12月20日 715 阅读
Abstract
Endophytic actinomycetes, particularly Streptomyces species, have gained significant attention due to their potential to produce novel bioactive compounds. In this study, we isolated and characterized an endophytic Streptomyces sp. VITGV100 from the tomato plant (Lycopersicon esculentum), employing the direct streak method and whole-genome sequencing. A genome analysis was done to uncover its biosynthetic potential and identify indole-type compounds. The strain's secondary metabolite production was evaluated through GC–MS analysis, and its antimicrobial activity was tested against selected human pathogenic bacteria. Our protocol outlines a comprehensive approach, describing the isolation and extraction of metabolites and genome mining for indole-type compounds. This isolate has potential pharmaceutical applications, accelerating the discovery of novel indole-type bioactive compounds.
Key features
• A systematic approach for isolating Streptomyces sp. VITGV100 from Lycopersicon esculentum, including surface sterilization and selective culturing on ISP2 medium.
• Whole-genome sequencing and annotation: High-throughput Illumina sequencing followed by genome assembly, annotation, and functional characterization to explore biosynthetic potential and metabolic pathways.
• Secondary metabolite profiling and bioactivity screening: GC–MS-based identification of bioactive metabolites, followed by antibacterial activity assessment against human pathogens using well-diffusion assays.
• Identification of biosynthetic gene clusters (BGCs) using antiSMASH 6.0.
Keywords: Endophytic Streptomyces (内生链霉菌)Graphical overview
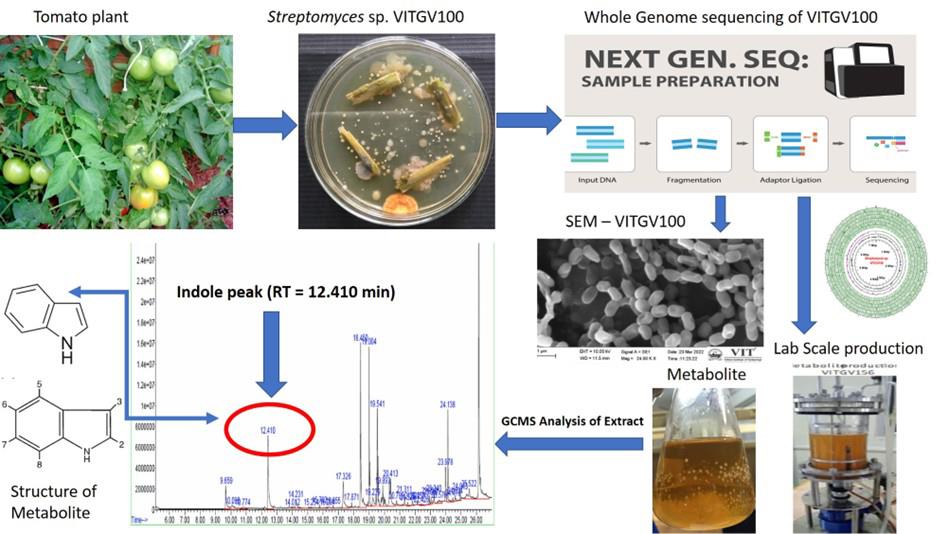
Genomic and metabolomic insights into the isolation of endophytic Streptomyces sp. VITGV100: a potential source of bioactive indole compounds
Background
Actinomycetes, particularly those belonging to the genus Streptomyces, are a prolific source of bioactive secondary metabolites with significant applications in medicine, agriculture, and industry. Among them, Streptomyces species have garnered attention for their potential to produce novel antimicrobial, anticancer, and even plant growth-promoting compounds [1]. Microbial strains offer a sustainable and environmentally friendly alternative to synthetic agrochemicals, playing a crucial role in biocontrol and plant health enhancement [2]. Traditional methods for isolating Streptomyces from environmental sources involve selective culturing techniques using specific growth media, followed by morphological and biochemical characterization [3]. However, recent advances in molecular biology and whole genome sequencing (WGS) have revolutionized the identification and functional analysis of these microorganisms [4]. WGS enables comprehensive insights into the genetic architecture of Streptomyces, facilitating the discovery of biosynthetic gene clusters (BGCs) responsible for secondary metabolite production [5]. This genomic approach overcomes the limitations of conventional screening methods, which often fail to detect cryptic or silent gene products under standard laboratory conditions [6]. The protocol described herein focuses on the isolation, genome sequencing, and functional characterization of Streptomyces sp. VITGV100, an endophytic actinomycete strain isolated from Lycopersicon esculentum (tomato) plants [5]. Compared to traditional bioactivity-guided screening, this genomic approach enhances the efficiency of novel bioactive compound discovery by directly linking genetic information to metabolite production [7]. A key advantage of this protocol is its ability to systematically identify and analyze BGCs involved in secondary metabolite biosynthesis using tools such as antiSMASH [8] and ARTS [9]. This computational approach significantly accelerates the identification of novel antibiotics [10]. Furthermore, comparative genomic analyses using ANI and dDDH facilitate the taxonomic positioning and evolutionary relationships of the newly isolated strain, contributing to a deeper understanding of Streptomyces diversity and genetic variability [11]. Beyond antibiotic discovery, the protocol has broad applications in biotechnology and agriculture. Endophytic Streptomyces strains have been reported to enhance plant growth through phytohormone production, nitrogen fixation, and antagonism against phytopathogens [12]. In summary, this protocol represents a comprehensive strategy for isolating, characterizing, and harnessing the biosynthetic potential of endophytic Streptomyces strains.
Materials and reagents
Biological materials
1. Healthy 4-week-old tomato plants grown in natural soil under conditions (Lycopersicon esculentum) (Arka Rakshak)
2. Streptomyces sp. VITGV100 (MCC 4961)
4. Staphylococcus aureus (MTCC 737)
5. Escherichia coli (MTCC 1687)
Reagents
1. International Streptomyces Project -2 agar (Himedia, CAS number: 8002-48-0)
2. Tetracycline (Himedia, CAS number: 64-75-5)
3. Cetyltrimethylammonium bromide (Himedia, AS number: 57-09-0)
4. Cyclohexamide (Himedia, CAS number: 66-81-9)
5. Nystatin (Himedia, CAS number: 1400-61-9)
6. Streptomyces DNA Purification kit (Himedia, CAS number: MBA527)
7. Ethanol (Himedia, CAS number: 64-17-5)
8. NaHCO3 (Himedia, CAS number: 144-55-8)
9. Ethyl acetate (Himedia, CAS number: 141-78-6)
10. Methanol (Himedia, CAS number: 67-56-1)
11. Glycerol (Himedia, CAS number: 56-81-5)
12. Sodium hypochlorite (Himedia, CAS number: 7681-52-9)
13. Mueller Hinton agar (MHA) (Himedia, CAS number: M1084)
Solutions
1. Yeast malt agar - ISP2 agar (see Recipes)
2. MHA agar (see Recipes)
Recipes
1. Yeast malt agar - ISP2 agar
| Ingredients | Final concentration |
|---|---|
| Peptone | 5 g/L |
| Yeast extract | 3 g/L |
| Malt extract | 3 g/L |
| Dextrose (glucose) | 10 g/L |
| Agar | 20 g/L |
2. MHA agar
| Ingredients | Final concentration |
|---|---|
| HM infusion solids B # (from 300 g) | 2 g/L |
| Acicase | 17.5 g/L |
| Starch | 1.5 g/L |
| Agar | 17. g/L |
Laboratory supplies
1. Erlenmeyer flasks (Borosil Scientific, catalog number: 4980)
2. Separating funnel (Borosil Scientific, catalog number: 6403)
3. Round-bottom flask (Borosil Scientific, catalog number: 4380)
4. Storage bottle (Borosil Scientific, catalog number: 1519)
5. Sample vials (Borosil Scientific, catalog number: 9913)
6. Petri plates (Borosil Scientific, catalog number: 3165)
7. Cork borer (HiMedia, India: LA737)
Equipment
1. Incubator (Thermo Scientific, catalog number: 50125590)
2. Hot air oven (Thermo Scientific, catalog number: PR305225M)
3. Biological safety cabinet, HEPA filter (Thermo Scientific, catalog number: 51025411)
4. Rotary vacuum evaporator (Fisher Scientific, catalog number: 05-405-195)
5. Gas chromatography and mass spectrophotometry (Thermo Scientific, model: TOEXGFTH)
6. Nanodrop (Thermo Scientific, model: ND-2000C)
7. Centrifuge (Remi, model: R-24)
8. Cooling centrifuge (Remi, model: CM-12)
9. Zone measuring scale (HiMedia, India, model: PW297)
10. GC–MS (Thermo Scientific Trace GC Ultra & ISQ Single Quadrupole MS, USA)
Software and datasets
1. antiSMASH 6.0 (Manufacturer, Version - https://antismash.sondarymetabolites.org)
2. Alignment Search Tool (BlastX): https://blast.ncbi.nlm.nih.gov/Blast.cgi
3. Kyoto Encyclopedia of Genes and Genomes (KEGG):
3. KEGG Automatic Annotation Server: https://www.genome.jp/tools/kaas/
4. CGView server 1.0: https://proksee.ca/
5. Blast2GO platform: https://www.blast2go.com
6. Web Gene Ontology Annotation plot (http://wego.genomics.org.cn/cgi-bin/wego/index.pl)
7. MegaX 6.0: https://www.megasoftware.net/
8. Antimicrobial Resistant Target Seeker: https://arts.ziemertlab.com
9. Prokka (version 1.12): https://github.com/tseemann/prokka
10. SPAdes assembler (v-3.13.0): https://github.com/ablab/spades
11. Trimmomatic v0.38: http://www.usadellab.org/cms/?page=trimmomatic
12. Streptomyces sp. VITGV100: https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR15294182&display=analysis
Procedure
文章信息
稿件历史记录
提交日期: Apr 11, 2025
接收日期: Jun 15, 2025
在线发布日期: Jul 4, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Pattapulavar, V., Ramanujam, S., Muthusamy, S., Panchal, S. and Christopher, J. G. (2025). Metabolite Production and Extraction of Indole Compound From the Tomato Endophyte Streptomyces sp. VITGV100. Bio-protocol 15(14): e5386. DOI: 10.21769/BioProtoc.5386.
分类
微生物学 > 微生物遗传学 > 全基因组测序
生物化学 > 其它化合物 > 小分子
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link