- EN - English
- CN - 中文
Evaluation of Translation Rate Through L-azidohomoalanine (AHA) Incorporation and Subsequent Alkyne Fluorophore–Mediated Click Chemistry in Yeast
酵母中蛋白质合成速率的评估:基于L-叠氮高半胱氨酸掺入与炔基荧光探针点击化学反应的方法
(§Technical contact: mj216@snu.edu.in) 发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5379 浏览次数: 2560
评审: Willy R Carrasquel-UrsulaezAnonymous reviewer(s)
Abstract
Accurate measurement of protein translation rates is crucial for understanding cellular processes and disease mechanisms. However, existing methods for quantifying translation rates in yeast cells are limited. Here, we present a streamlined protocol for measuring protein translation rates in Saccharomyces cerevisiae using the methionine analog L-azidohomoalanine (AHA), which is the L isoform of this synthetic amino acid, and fluorophore-labeled alkyne dye-based Click chemistry. Our method involves incorporating AHA into newly synthesized proteins, followed by detection using confocal microscopy, flow cytometry, and SDS-PAGE. We validated our protocol by measuring translation rates under various stress conditions, including heat stress, endoplasmic reticulum (ER) stress induced by tunicamycin, and translation inhibition by cycloheximide. Confocal microscopy revealed differential AHA incorporation and fluorescence intensity across conditions. Flow cytometry quantitatively confirmed significant increases in translation rates under heat stress and decreases under ER stress compared to unstressed conditions at 6 and 24 h post-treatment. Imaging of gels under fluorescence detectors following SDS-PAGE further visualized newly synthesized proteins, with no detectable translation after cycloheximide treatment. Our protocol offers enhanced precision and selectivity compared to existing methods for mammalian cells and represents the first standardized approach for measuring translation rates in yeast. Despite limitations in required specialized equipment and expertise, this method holds promise for diverse applications in biotechnology and biomedical research, enabling investigations into protein synthesis regulation in yeast systems.
Key features
• This study presents the first standardized protocol for measuring protein translation in budding yeast using AHA and Click chemistry, addressing yeast-specific challenges effectively.
• The study uses microscopy, flow cytometry, and fluorescence gel imaging to robustly validate yeast translation rates, ensuring reliable, reproducible results across cellular and biochemical levels.
• The method detects translation changes under stress: increased with heat, decreased with ER stress, and halted by cycloheximide, highlighting its sensitivity for proteostasis research.
• Despite requiring specialized equipment and expertise, the method offers valuable applications in biomedical research, metabolic engineering, and drug screening focused on protein homeostasis in yeast.
Keywords: Click chemistry (点击化学)Graphical overview
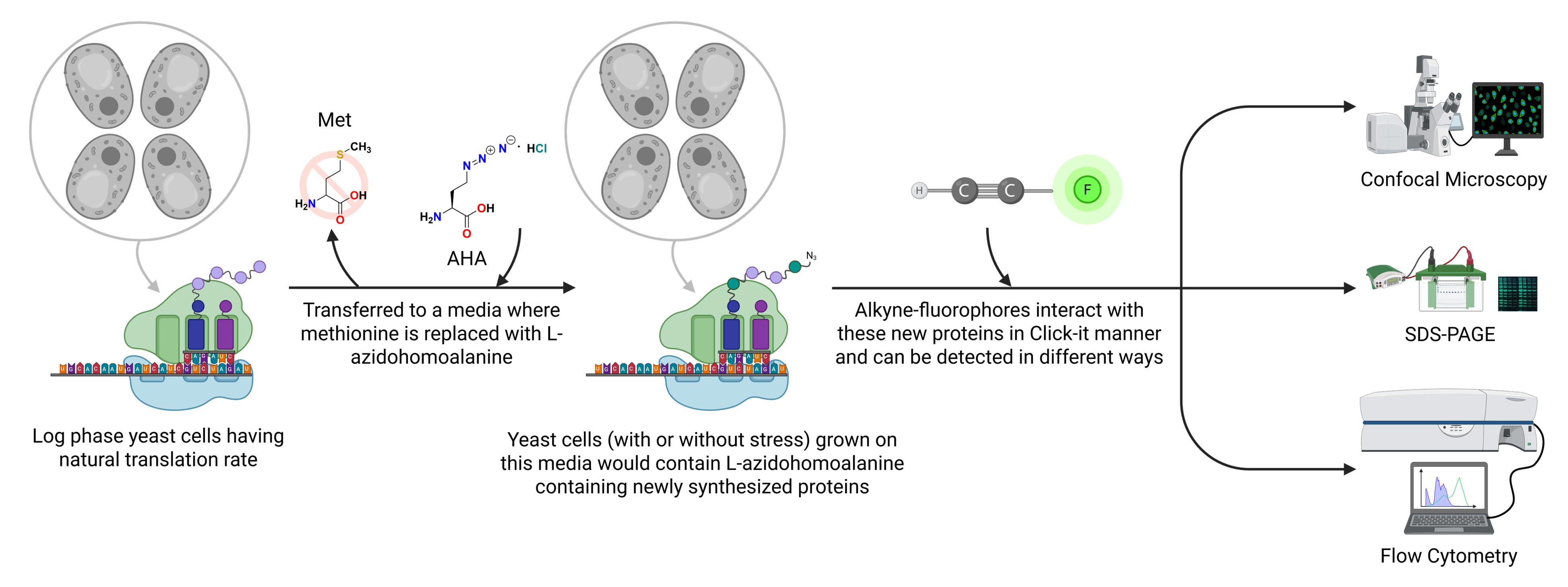
Schematic overview of the L-azidohomoalanine (AHA)-based Click chemistry protocol to measure translation rate in Saccharomyces cerevisiae. Schematic representation showing L-azidohomoalanine incorporated into newly synthesized proteins that can be detected by using alkyne-modified fluorophores and qualitatively and quantitatively measured by various techniques, as depicted in the figure.
Background
Protein translation is a critical cellular process that converts genetic information from DNA into functional proteins, which are essential for various cellular activities such as biochemical catalysis, structural integrity, and intercellular signaling. This process involves the conversion of messenger RNA (mRNA) into proteins through tightly regulated stages, including initiation, elongation, termination, and ribosome recycling, thereby ensuring accurate protein synthesis in response to internal and external stimuli [1]. Precision in protein production is vital for cellular function and organismal homeostasis, with aberrations potentially leading to dysfunctional proteins and pathological conditions [2].
Regulation of protein translation, which is crucial for cellular equilibrium and environmental adaptation, involves mRNA structure, translation factors, and aminoacyl-tRNA. The 5'-UTR of mRNA contains motifs that regulate translation stages, while secondary structures and sequences affect ribosomal access. Initiation factors assemble the translation complex, elongation factors add amino acids, and release factors terminate the synthesis. Aminoacyl-tRNAs deliver amino acids, affecting translation rate and efficiency [2]. Under stress conditions, cells store mRNA in stress granules and P-bodies [3]. Disrupted protein translation regulation is central to diseases such as cancer, neurodegenerative disorders, and metabolic disorders owing to altered translation factors, tRNA availability, or ribosome defects. Neurodegenerative diseases involve the accumulation of misfolded proteins. Advances in ribosome profiling and mass spectrometry have enhanced the study of translation errors and their impact on disease, suggesting new therapeutic approaches [4].
Ribosomes can simultaneously translate multiple mRNAs to form polysomes and enhance their efficiency. The movement of mRNA through the ribosome affects the speed of protein synthesis, with suboptimal mRNA structures hindering ribosome binding and movement, thus reducing efficiency [3]. Translation rates depend on the translational machinery and factors such as nutrient availability and stress responses. Nutrient abundance supplies essential components for protein synthesis, whereas nutrient scarcity can halt translation. Stress can sequester mRNA in stress granules, reducing the available mRNAs for translation and conserving energy. During stress, translation regulation enables cells to balance protein synthesis with immediate needs, prioritize critical functions, and optimize survival in changing environments [2,5].
This study presents a streamlined methodology for accurately measuring the protein translation rate in the budding yeast Saccharomyces cerevisiae using a methionine analog, L-azidohomoalanine (AHA), and subsequent fluorophore-labeled alkyne dye-based Click chemistry reaction (Figure 1). While Click chemistry–based detection techniques are available for mammalian cells, none address the complexities associated with budding yeast samples. Our protocol delineates the detailed step-by-step process of determining the protein translation rate in budding yeast; our results were validated by incorporating L-azidohomoalanine into newly synthesized proteins using flow cytometry, microscopy, and visualization in protein gels. In essence, our protocol effectively establishes a convenient method for evaluating protein translation rate in budding yeast under various growth conditions.
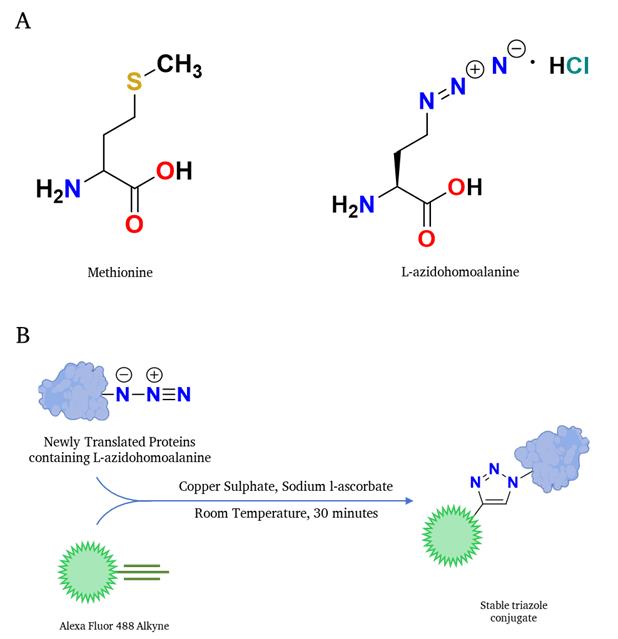
Figure 1. Schematic representation of the translation rate determination through Click chemistry–based reaction. (A) Structure of methionine is shown on the left, and the structure of the methionine analog L-azidohomoalanine is shown on the right. (B) L-azidohomoalanine, incorporating newly synthesized proteins, interacts with the alkyne-containing fluorophore in a Click-IT reaction fashion; subsequently, the fluorophore-labeled newly synthesized proteins can be detected in any fluorescence-detection system.
Materials and reagents
Biological materials
1. Yeast strain BY4741 (genotype: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; parent strain: S288C; source: S288C/EUROSCARF)
Reagents
1. Glucose (Sigma, catalog number: G8270)
2. Yeast nitrogen base (Himedia, catalog number: M151)
3. Ammonium chloride (MP Biomedicals, catalog number: 150109)
4. Methionine (Sigma, catalog number: M9625)
5. Phenylalanine (Sigma, catalog number: P5482)
6. Adenine (Sigma, catalog number: A2786)
7. Arginine (Sigma, catalog number: A8094)
8. Threonine (Sigma, catalog number: T8441)
9. Lysine (Sigma, catalog number: L9037)
10. L-azidohomoalanine (Sigma, catalog number: 900892)
11. Histidine (Sigma, catalog number: H5659)
12. Leucine (Sigma, catalog number: L8912)
13. Tryptophan (Sigma, catalog number: T8941)
14. Uracil (Sigma, catalog number: U1128)
15. Agar powder, bacteriological grade (Himedia, catalog number: GRM026)
16. Tunicamycin (Sigma, catalog number: T7765)
Note: Light sensitive, store at -20 °C, prepare dilutions right before use as a stressor in cultures.
17. Cycloheximide (Sigma, catalog number: C4859)
Note: Light sensitive, store at -20 °C, prepare dilutions right before use as a stressor in cultures.
18. Sodium hydroxide (MP Biomedicals, catalog number: 153495)
19. Sodium chloride (MP Biomedicals, catalog number: 102892)
20. Potassium chloride (MP Biomedicals, catalog number: 151944)
21. Sodium phosphate dibasic (MP Biomedicals, catalog number: 191440)
22. Potassium phosphate dibasic (MP Biomedicals, catalog number: 191431)
23. Hydrochloric acid (Sigma, catalog number: H9892)
24. Tris (MP Biomedicals, catalog number: 194855)
25. EDTA disodium dihydrate (MP Biomedicals, catalog number: 194822)
26. Phenylmethylsulfonyl fluoride (PMSF) (MP Biomedicals, catalog number: 195381)
27. Sodium dodecyl sulfate (MP Biomedicals, catalog number: 811033)
28. Glycerol (MP Biomedicals, catalog number: 193996)
29. Bromophenol blue (MP Biomedicals, catalog number: 805732)
30. β-Mercaptoethanol (MP Biomedicals, catalog number: 190242)
31. Acid-washed glass beads (Sigma, catalog number: G8772)
32. Molecular biology grade absolute ethanol (Hayman premium grade, 100%, catalog number: F205855)
33. Alexa-Fluor 488 alkyne (ThermoFisher, catalog number: A10267)
Note: Light sensitive, store at -20 °C, prepare dilutions right before use as a Click reagent for permeabilized cells.
34. Copper (II) sulphate pentahydrate (Sigma, catalog number: C8027)
35. Sodium L-ascorbate (Sigma, catalog number: 11140)
36. Liqui-gel, premixed acrylamide/bis-acrylamide solution (MP Biomedicals, catalog number: 800803)
Solutions
1. Synthetic complete media (see Recipes)
2. Synthetic complete media with L-azidohomoalanine (see Recipes)
3. PBS 10× stock (see Recipes)
4. Yeast permeabilization buffer (see Recipes)
5. Click reaction buffer (see Recipes)
6. Sodium hydroxide solution 0.3 M (see Recipes)
7. Protein storage buffer (see Recipes)
8. SDS loading dye 5× stock (see Recipes)
Recipes
1. Synthetic complete media
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Glucose | 20 mg/mL | 10 mL of 200 mg/mL stock |
| Yeast nitrogen base | 1.7 mg/mL | 10 mL of 17 mg/mL stock |
| Ammonium chloride | 5 mg/mL | 10 mL of 50 mg/mL stock |
| Methionine | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Phenylalanine | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Adenine | 0.03 mg/mL | 1 mL of 3 mg/mL stock |
| Arginine | 0.03 mg/mL | 1 mL of 3 mg/mL stock |
| Threonine | 0.15 mg/mL | 1 mL of 15 mg/mL stock |
| Lysine | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Histidine | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Leucine | 0.1 mg/mL | 1 mL of 10 mg/mL stock |
| Tryptophan | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Uracil | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
2. Synthetic complete media with L-azidohomoalanine
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Glucose | 20 mg/mL | 10 mL of 200 mg/mL stock |
| Yeast nitrogen base | 1.7 mg/mL | 10 mL of 17 mg/mL stock |
| Ammonium chloride | 5 mg/mL | 10 mL of 50 mg/mL stock |
| L-azidohomoalanine | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Phenylalanine | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Adenine | 0.03 mg/mL | 1 mL of 3 mg/mL stock |
| Arginine | 0.03 mg/mL | 1 mL of 3 mg/mL stock |
| Threonine | 0.15 mg/mL | 1 mL of 15 mg/mL stock |
| Lysine | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Histidine | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Leucine | 0.1 mg/mL | 1 mL of 10 mg/mL stock |
| Tryptophan | 0.05 mg/mL | 1 mL of 5 mg/mL stock |
| Uracil | 0.02 mg/mL | 1 mL of 2 mg/mL stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
3. PBS 10× stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sodium chloride | 80 mg/mL | 8,000 mg |
| Potassium chloride | 2 mg/mL | 200 mg |
| Sodium phosphate dibasic | 17.8 mg/mL | 1,780 mg |
| Potassium phosphate dibasic | 2.4 mg/mL | 240 mg |
| Hydrochloric acid | to adjust pH to 7.4 | As required from 100% stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
Note: Dilute to 1× with sterile distilled water as needed.
4. Yeast permeabilization buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Absolute ethanol | 53% | 53 mL of 100% stock |
| PBS | 1× | 10 mL of 10× stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
5. Click reaction buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-Cl | 2,000 mM | 5 mL of 4 M pH 8.5 stock |
| Copper sulphate | 50 mM | 0.5 mL of 1 M stock |
| Alexa-Fluor 488 alkyne | 1 μg/mL | 10 μL of 1 mg/mL stock |
| Ascorbic acid | 500 mM | 1.667 mL of 3 M stock |
| Autoclaved sterile water | n/a | Up to 10 mL |
| Total | n/a | 10 mL |
6. Sodium hydroxide solution 0.3 M
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sodium hydroxide | 300 mM | 1,200 mg |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
7. Protein storage buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-Cl | 10 mM | 1 mL of 1 M pH 7.4 stock |
| EDTA | 1 mM | 0.2 mL of 0.5 M pH 8 stock |
| Potassium chloride | 150 mM | 3.75 mL of 4 M stock |
| PMSF | 1 mM | 0.5 mL of 200 mM stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
8. SDS loading dye 5× stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-Cl | 250 mM | 25 mL of 1 M pH 6.8 stock |
| Sodium dodecyl sulfate | 4% | 20 mL of 20% stock |
| Glycerol | 30% | 30 mL of 100% stock |
| Bromophenol blue | 0.6 mg/mL | 60 mg |
| β-Mercaptoethanol | 16% | 16 mL of 100% stock |
| Autoclaved sterile water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
Laboratory supplies
1. Micropipette tips: 10 μL, 100 μL, and 1,000 μL (Tarsons, catalog numbers: 521010, 521010, and 521020, respectively)
2. Petri dishes (Tarsons, catalog number: 460030)
3. 1.5 mL microcentrifuge tubes (Tarsons, catalog number: 500010)
4. 50 mL centrifuge tubes (Tarsons, catalog number: 546041)
5. Screw-capped glass flasks of 25 and 100 mL (Borosil, catalog numbers: 1501009 and 5021016, respectively)
6. Single-channel pipets, 0.2–2 μL (P2), 2–20 μL (P20), 20–200 μL (P200), 100–1,000 μL (P1000) (Gilson, PIPETMANS, catalog numbers: F144054M, F144056M, F144058M, and F144059M, respectively)
Equipment
1. Autoclave (ALP, catalog number: 32SDP)
2. Shaker incubator (Scigenics Biotech Orbitek, catalog number: LE-BT-AH)
3. Spectrophotometer (Biospectrometer, Eppendorf, catalog number: 6136000.002)
4. Centrifuges for 50 mL tubes and 1.5 mL microcentrifuge tubes (Eppendorf 5810R and 5425R, catalog numbers: EP022627066 and EP5406000046, respectively)
5. Bead-Ruptor (Omni International, catalog number: 25-010)
6. Laser scanning confocal microscope (Nikon, model: A1R MP+ Multiphoton Advanced Resolution)
7. Fluorescence activated cytometer (Beckman Coulter, model: Cytoflex S)
8. SDS-PAGE unit (Bio-Rad, model: Mini-PROTEAN Tetra Vertical Electrophoresis Cell)
9. Fluorescence-enabled Gel Doc system (Invitrogen, model: iBright)
10. Gel Doc system (Bio-Rad, model: Gel Doc EZ imager, catalog number: 170-8270)
Software and datasets
1. Nikon NIS-Elements AR (Nikon, 5.20.02); available with the microscope; otherwise, requires a license
2. CytExpert (Beckman Coulter, 2.5.0.77); available with the fluorescence analyzer; otherwise, requires a license
3. FlowJo (FlowJo LLC, 10.8); requires a license
4. GraphPad Prism (GraphPad Software, 9.5.0); requires a license
5. Biorender portal (Biorender, https://app.biorender.com/); requires a license
6. MS Office suite, Excel, PowerPoint, and Word (Microsoft, Office 365); requires a license
Procedure
文章信息
稿件历史记录
提交日期: Apr 17, 2025
接收日期: Jun 10, 2025
在线发布日期: Jul 3, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Jha, M. P. and Mapa, K. (2025). Evaluation of Translation Rate Through L-azidohomoalanine (AHA) Incorporation and Subsequent Alkyne Fluorophore–Mediated Click Chemistry in Yeast. Bio-protocol 15(14): e5379. DOI: 10.21769/BioProtoc.5379.
分类
分子生物学 > 蛋白质 > 检测
生物化学 > 蛋白质 > 标记
细胞生物学 > 基于细胞的分析方法 > 蛋白合成
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













