- EN - English
- CN - 中文
APEX2 RNA Proximity Labeling in Mammalian Cell Lines With Low Biotin Permeability
在低生物素通透性的哺乳动物细胞系中进行APEX2介导的RNA邻位标记
发布: 2025年07月05日第15卷第13期 DOI: 10.21769/BioProtoc.5372 浏览次数: 2346
评审: Marion HoggAnik TuladharAnonymous reviewer(s)
Abstract
The subcellular localization of RNA plays a critical role in various biological processes, including development and stress response. Proximity labeling eases the detection of localized transcripts and protein enrichment compared to previous techniques that rely on biochemical isolation of subcellular structures. The rapid reaction and small labeling radius of APEX2 make it an attractive alternative to other proximity labeling approaches, such as BioID. However, we found that standard protocols for APEX proximity labeling fail in human induced pluripotent stem cells. Moreover, standard protocols yield heterogeneous labeling of biomolecules across single cells in MCF10A breast epithelial cells. Our results indicate that low biotin permeability in these cell lines is the main cause for failed or inefficient labeling. This protocol outlines improved labeling by combining the rapid hydrogen peroxide-driven APEX2 reaction with the addition of a mild detergent during biotin incubation. This adaptation leads to efficient proximity labeling in hiPSCs and more homogeneous biotinylation across single cells in MCF10As. The adapted protocol extends the use of APEX2 proximity labeling to cell lines with poor biotin permeability.
Key features
• Builds on methods developed by the Ting Lab [1] and the Ingolia Lab [2] for proximity labeling of transcripts using the enzyme APEX2.
• Focuses on cell lines with low biotin permeability, like human induced pluripotent stem cells, by including a mild detergent to increase biotin uptake.
• Includes controls for nonspecific molecular localization and statistical methods for processing of resulting sequencing data.
Keywords: RNA proximity labeling (RNA邻位标记)Graphical overview
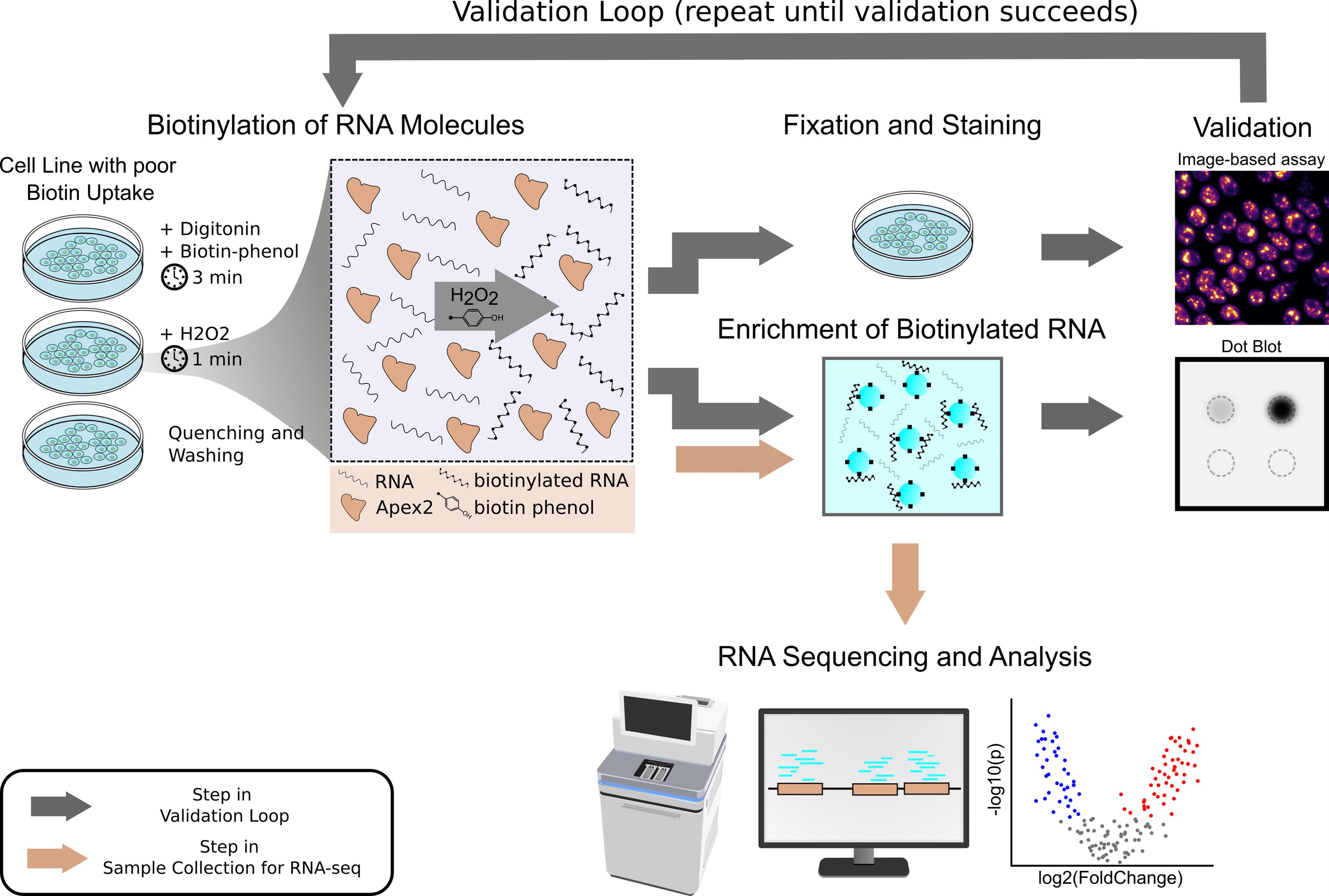
Workflow for proximity labeling of subcellular RNA and enrichment of biotinylated RNA in human induced pluripotent stem cells and other cell lines with low biotin permeability
Background
Within cells, both direct binding between molecules and weaker, more transient molecular interactions regulate cellular activity. Proximity labeling approaches have emerged as a way to capture spatial information within cells and to identify potential interactions among proteins and nucleic acids independent of stable binding. Several methods exist to biotinylate and pull-down biomolecules based on proximity to a bait protein, which differ in the enzyme used for biotinylation and in reaction kinetics [3]. Whether the enzyme is a peroxidase that requires H2O2for activation, like horseradish peroxidase or ascorbate peroxidase 2 (APEX2), or a biotin ligase, as in BioID or TurboID, conjugating the biotinylating enzyme to a bait protein of interest allows pull-down of nearby biomolecules using streptavidin-coated beads following biotinylation. Both proteins and nucleic acids undergo biotinylation, enabling the subsequent identification of biotinylated proteins through mass spectrometry analysis, or DNA or RNA through sequencing. Compared to other methods, APEX2 offers a short and efficient reaction, although dependence on H2O2 restricts in vivo applications.
APEX2 proximity labeling has proven useful in identifying the components of phase-separated compartments and other subcellular organelles, particularly membrane-associated regions [1,2,4,5]. While most APEX2 studies have used cell lines that readily take up exogenous biotin, we found that standard protocols fail to achieve efficient biotinylation in human induced pluripotent stem cells (hiPSCs) and produce heterogeneous labeling in MCF10A breast epithelial cells. Adapting published APEX2 protocols to these two cell lines with poor biotin permeability required optimizing the addition of a mild detergent to allow biotin entry into the cells. Unlike other proximity labeling approaches that require long reaction times and, therefore, long incubation periods with biotin, the temporal resolution of APEX2 is compatible with a quick application of detergent followed by cell harvesting. Here, we present an example application for our adapted APEX2 biotin labeling protocol in hiPSCs endogenously expressing the APEX2-tagged protein SON, a scaffold protein of nuclear speckles, to identify RNAs enriched in this compartment. We use an endogenous tagging strategy in hiPSCs to avoid possible artifacts introduced by overexpression of APEX2 constructs and allow for future differentiation for comparison of RNA localization across cell types. However, this protocol also extends to other mammalian cell lines, including MCF10As, and to proximity labeling using transiently or stably transfected APEX2 constructs. Similar steps prior to RNA extraction could also be adapted for proteomics analysis of biomolecular compartments in these cell lines.
Materials and reagents
Biological materials
1. Cell line expressing a protein of interest tagged with the APEX2 enzyme. For example, WTC-11 hiPSCs carrying an endogenous APEX2-GFP tag (~59 kDa) at the SON locus for nuclear speckle tagging (referred to as APEX2-SON hiPSCs, generated in-house)
2. Control cell line expressing the APEX2 enzyme with nonspecific molecular localization. For example, WTC-11 hiPSCs expressing APEX2-GFP-NLS within a safe-harbor locus for general nuclear tagging (referred to as APEX2-NLS hiPSCs, generated in-house)
Note: To generate the endogenously tagged hiPSC lines, we followed protocols from the Allen Institute for Cell Sciences [6,7]
Reagents
1. (+)-Sodium L-ascorbate (Sigma-Aldrich, catalog number: A7631)
2. 16% paraformaldehyde (Electron Microscopy Sciences, catalog number: 15710)
3. Biotin-phenol (Iris Biotech, catalog number: 41994-02-9)
4. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: B4287-5G)
5. Digitonin (Sigma-Aldrich, catalog number: D141)
6. Dithiothreitol (DTT) (Thermo Fisher Scientific, catalog number: D1532)
7. DPBS (Thermo Fisher Scientific, catalog number: 14190250)
8. EDTA (0.5 M), pH 8.0, RNase-free (Thermo Fisher Scientific, catalog number: AM9260)
9. Hydrogen peroxide solution (30%) (Sigma-Aldrich, catalog number: H1009-5ML)
10. InvitrogenTM NaCl (5 M), RNase-free (Thermo Fisher Scientific, catalog number: AM9760G)
11. NaCl (5 M), RNase-free (Thermo Fisher Scientific, catalog number: AM9760G)
12. N-Lauroylsarcosine sodium salt solution (Sigma-Aldrich, catalog number: L7414)
13. Paraformaldehyde 16% aqueous solution EM grade (EMS, catalog number: 15600)
14. Phosphate-buffered saline (10×) pH 7.4, RNase-free (Thermo Fisher Scientific, catalog number: AM9625)
15. RNase A Solution (Sigma-Aldrich, catalog number: 70856-3)
16. RNaseOUTTM recombinant ribonuclease inhibitor (Thermo Fisher Scientific, catalog number: 10777019)
17. Sodium azide (Merck, catalog number: s2002)
18. Sodium hydroxide solution (Sigma-Aldrich, catalog number: 72068)
19. Streptavidin, Alexa FluorTM 647 conjugate (Thermo Fisher Scientific, catalog number: S21374)
20. SUPERase·InTM RNase inhibitor (20 U/μL) (Thermo Fisher Scientific, catalog number: AM2696)
21. TritonTM X-100 Surfact-AmpsTM detergent solution (Thermo Fisher Scientific, catalog number: 85112)
22. Trolox (Sigma-Aldrich, catalog number: 238813)
23. TWEEN® 20 (Sigma-Aldrich, catalog number: P9416)
24. UltraPureTM 1 M Tris-HCI buffer, pH 7.5 (Thermo Fisher Scientific, catalog number: 15567027)
25. UltraPureTM DNase/RNase-free distilled water, 500 mL (Thermo Fisher Scientific, catalog number: 10977015)
Solutions
1. Biotin-phenol stock solution (see Recipes)
2. Digitonin solution (5%) (see Recipes)
3. H2O2 stock solution (see Recipes)
4. Trolox stock solution (see Recipes)
5. Sodium azide stock solution (see Recipes)
6. Sodium ascorbate stock solution (see Recipes)
7. Biotin-phenol labeling solution (see Recipes)
8. Quenching solution (see Recipes)
9. Washing solution (see Recipes)
10. 3× Proteinase buffer (see Recipes)
11. 2× Fixation solution (see Recipes)
12. 2× Permeabilization solution (see Recipes)
13. 2× Blocking solution (see Recipes)
14. Fluorescent streptavidin solution (see Recipes)
15. Binding and washing buffer (see Recipes)
16. Solution A (see Recipes)
17. Solution B (see Recipes)
Recipes
1. Biotin-phenol stock solution
Note: Prepare aliquots and store at -80 °C.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Biotin-phenol | 250 mM | 90.8675 mg |
| DMSO | n/a | 926 μL |
| Total | n/a | 1 mL |
2. Digitonin solution (5%)
Note: The solution has to be heated to 95–98 °C to fully dissolve. The solution can be stored at room temperature but will form precipitates. Prewarm the solution until it is clear before use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Digitonin | 5% | 100 mg |
| H2O (RNase-free) | n/a | 2 mL |
| Total | n/a | 2 mL |
3. H2O2 stock solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Hydrogen peroxide solution (30%) | 100 mM | 10 μL |
| H2O (RNase-free) | n/a | 1 mL |
| Total | n/a | 1 mL |
4. Trolox stock solution
Note: Prepare Trolox stock solution freshly before use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Trolox | 500 mM | 31.3 mg |
| DMSO | n/a | 226 μL |
| Total | n/a | 250 μL |
5. Sodium azide stock solution
Note: Sodium azide stock solution can be stored at room temperature.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sodium azide | 2% | 200 mg |
| H2O (RNase-free) | n/a | 10 mL |
| Total | n/a | 10 mL |
6. Sodium ascorbate stock solution
Note: Prepare sodium ascorbate stock solution freshly before use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sodium ascorbate | 1 M | 100 mg |
| H2O (RNase-free) | n/a | 440 μL |
| Total | n/a | 500 μL |
7. Biotin-phenol labeling solution
Note: Prepare biotin-phenol labeling solution freshly before use. The addition of digitonin might not be necessary for all cell lines. If biotin take-up is efficient, the addition of digitonin can be omitted.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Biotin-phenol stock solution | 0.5 mM | 10 μL |
| Digitonin solution (5%) | 0.005% | 10 μL |
| SUPERase·InTM RNase inhibitor | 1:1000 | 10 μL |
| DPBS (1×) | n/a | 9.97 mL |
| Total | n/a | 10 mL |
8. Quenching solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Trolox (500 mM stock) | 5 mM | 100 μL |
| Sodium ascorbate stock solution | 10 mM | 100 μL |
| Sodium azide stock solution | 10 mM | 320 μL |
| SUPERase·InTM RNase inhibitor | 1:1,000 | 10 μL |
| DPBS (1×) | n/a | 9.47 mL |
| Total | n/a | 10 mL |
9. Washing solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Trolox stock solution | 5 mM | 300 μL |
| Sodium ascorbate stock solution | 10 mM | 300 μL |
| SUPERase·InTM RNase inhibitor | 1:1,000 | 30 μL |
| DPBS (1×) | n/a | 29.37 mL |
| Total | n/a | 30 mL |
10. 3× Proteinase buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PBS (10×) | 3× | 150 μL |
| N-Lauryl sarcosine sodium solution (20%) | 6% | 150 μL |
| EDTA (0.5 M) | 30 mM | 30 μL |
| DTT (1 M) | 15 mM | 7.5 μL |
| H2O (RNase-free) | n/a | 162.5 μL |
| Total | n/a | 500 μL |
11. 2× Fixation solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Paraformaldehyde 16% | 8% | 10 mL |
| PBS | n/a | 10 mL |
| Total | n/a | 20 mL |
12. 2× Permeabilization solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Triton X-100 (10%) | 0.5% | 500 μL |
| PBS | n/a | 9.5 mL |
| Total | n/a | 10 mL |
13. 2× Blocking solution
Note: Dilute 4 g of BSA in ~40 mL of PBS and place on a rotator. Adjust the volume to 50 mL once the BSA is fully dissolved.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| BSA | 4% | 4 g |
| PBS | n/a | ~50 mL |
| Total | n/a | 50 mL |
14. Fluorescent streptavidin solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Streptavidin, Alexa FluorTM 647 Conjugate | 1:1,000 | 1 μL |
| 2× blocking solution | n/a | 1 mL |
| Total | n/a | 1 mL |
15. Binding and washing buffer
Note: Binding and washing buffer can be prepared in advance and stored at room temperature.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 5 mM | 250 μL |
| EDTA (0.5 M) | 0.5 mM | 50 μL |
| NaCl (5 M) | 1 M | 10 mL |
| Tween-20 (100%) | 0.1% | 50 μL |
| H2O (RNase-free) | n/a | 39.65 mL |
| Total | n/a | 50 mL |
16. Solution A
Note: Solution A can be prepared in advance and stored at room temperature.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| NaOH (10 M) | 5 mM | 25 μL |
| NaCl (5 M) | 50 mM | 0.5 mL |
| H2O (RNase-free) | n/a | 49.475 mL |
| Total | n/a | 50 mL |
17. Solution B
Note: Solution B can be prepared in advance and stored at room temperature.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| NaCl (5 M) | 100 mM | 1 mL |
| H2O (RNase-free) | n/a | 49 mL |
| Total | n/a | 50 mL |
Laboratory supplies
1. 96-well imaging plate (Greiner, catalog number: 655096)
2. Chemiluminescent Nucleic Acid Detection Module kit (Thermo Fisher Scientific, catalog number: 89880)
3. DynabeadsTM MyOneTM Streptavidin C1 (Thermo Fisher Scientific, catalog number: 65001)
4. Eppendorf® DNA LoBind tubes (Eppendorf, catalog number: 0030108035)
5. High-sensitivity RNA ScreenTape (Agilent, catalog number: 5067-5579)
6. High-sensitivity RNA ScreenTape ladder (Agilent, catalog number: 5067-5581)
7. High-sensitivity RNA ScreenTape sample buffer (Agilent, catalog number: 5067-5580)
8. Hybond®-N+ hybridization membrane (Sigma-Aldrich, catalog number: GERPN203B)
9. QubitTM RNA High Sensitivity (HS) Assay kit (Thermo Fisher Scientific, catalog number: Q32852)
10. Quick-RNA Miniprep kit (Zymo Research, catalog number: R1054)
11. RNA Clean & Concentrator-5 (Zymo Research, catalog number: R1013)
12. SMARTer® Stranded Total RNA-Seq kit v2 - Pico Input Mammalian (Takara Bio, catalog number: 634411)
Equipment
1. iBrightTM CL750 Imaging System (Thermo Fisher Scientific, catalog number: A44116)
2. FisherbrandTM UV Crosslinker (Fisher Scientific, catalog number: 13-245-222)
3. Spinning disk confocal microscope (e.g., Visitron, model: VisiScope CSU-X1)
4. Optional: washer dispenser (Agilent, catalog number: EL406)
5. QubitTM fluorometer (Thermo Fisher Scientific, catalog number: Q33238)
6. TapeStation 4200 (Agilent, model: G2991BA)
Software and datasets
1. GENCODE gene annotations ≥ v40 [8]
2. R ≥ v.4.4.0 [9]
3. STAR ≥ v2.7.3a [10]
4. Rsubread ≥ v2.10.5 [11]
5. Trimmomatic ≥ v0.39 [12]
6. DESeq2 ≥ v1.36.0 [13]
Procedure
文章信息
稿件历史记录
提交日期: Apr 10, 2025
接收日期: Jun 4, 2025
在线发布日期: Jun 23, 2025
出版日期: Jul 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Tschan, A. B., Rai, A. K., Pelkmans, L. and McIntyre, A. B.R. (2025). APEX2 RNA Proximity Labeling in Mammalian Cell Lines With Low Biotin Permeability. Bio-protocol 15(13): e5372. DOI: 10.21769/BioProtoc.5372.
分类
分子生物学 > RNA > RNA 标记
细胞生物学 > 基于细胞的分析方法
生物化学 > RNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













