- 提交稿件
- 订阅
- CN
- EN - English
- CN - 中文
- EN - English
- CN - 中文
Untargeted Metabolomics of Epimastigote Forms of Trypanosoma cruzi
克氏锥虫表鞭毛体的非靶向代谢组学分析
发布: 2025年07月05日第15卷第13期 DOI: 10.21769/BioProtoc.5368 浏览次数: 807
评审: Anonymous reviewer(s)

相关实验方案

利用改进的培养方法和生长监测对嗜盐火山生菌进行表型分析的次氯酸盐胁迫试验
Paula Mondragon [...] Julie A. Maupin-Furlow
2022年11月20日 828 阅读
Abstract
Trypanosoma cruzi, the causative agent of Chagas disease, faces significant metabolic challenges due to fluctuating nutrient availability and oxidative stress within its insect vector. Metabolomic techniques, such as gas chromatography–mass spectrometry (GC–MS), have been widely used to study the adaptive mechanisms of the parasite. This article describes a standardized method for the untargeted metabolomics analysis of T. cruzi epimastigote, covering parasite cultivation, sample deproteinization with methanol, metabolite extraction, derivatization with BSTFA, and GC–MS analysis. To ensure robustness and reproducibility, statistical analysis uses univariate tests, as well as multivariate approaches such as principal component analysis (PCA) and partial least squares (PLS) regression. The protocol offers a reliable and sensitive method to study metabolic responses in T. cruzi under environmental stress, with low biological variability and high reproducibility.
Key features
• GC–MS was used to conduct a standardized metabolomics investigation of Trypanosoma cruzi epimastigote, assuring reproducibility and minimum biological variability.
• Includes sample deproteinization, metabolite extraction, and derivatization with BSTFA for accurate metabolite profiling under different biological conditions.
• Employs robust statistical approaches (PCA, PLS) to investigate differences among experimental groups and detect significant alterations in metabolism.
• Internal standards and multiple replicates ensure high sensitivity and repeatability, which is excellent for investigating metabolic processes in protozoan parasites.
Keywords: GC-MS (气相色谱-质谱)Graphical overview
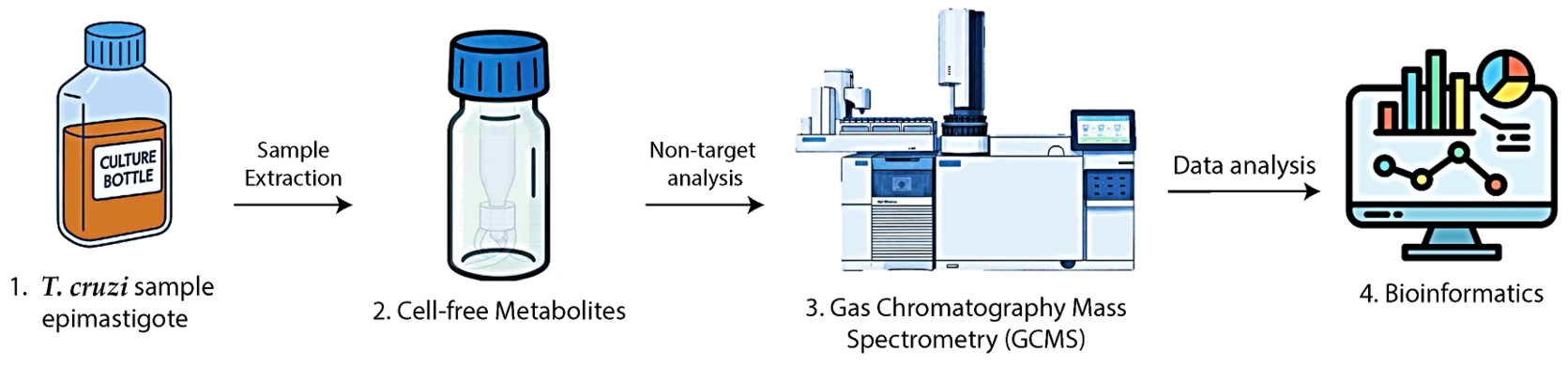 Graphical overview. 1. Trypanosoma cruzi epimastigotes are cultivated and counted. 2. Metabolite extracts are prepared, methoxylated, and silylated. 3. Samples are analyzed by GC–MS. 4. Data are processed using bioinformatics tools.
Graphical overview. 1. Trypanosoma cruzi epimastigotes are cultivated and counted. 2. Metabolite extracts are prepared, methoxylated, and silylated. 3. Samples are analyzed by GC–MS. 4. Data are processed using bioinformatics tools.
Background
Trypanosoma cruzi (T. cruzi) is a flagellated protozoan and the causative agent of Chagas disease, a neglected tropical disease affecting millions worldwide [1]. The parasite undergoes a complex life cycle, alternating between an insect vector and a mammalian host, which requires metabolic flexibility to survive fluctuating environmental conditions [2]. T. cruzi proliferates in the insect's midgut as epimastigotes, where it encounters nutrients obtained from the mammalian host's blood through feeding, generating oxidative products from red blood cell degradation. This environment exposes the parasite to reactive oxygen species (ROS) produced by hemoglobin breakdown, demanding adaptation mechanisms to handle oxidative stress and nutritional variations [3]. Understanding how the parasite responds to these stresses is critical for identifying potential metabolic limitations that can be targeted by therapeutic approaches for Chagas disease.
Metabolomic analyses have been used to explore T. cruzi's metabolic adaptations to various conditions [4]. This and other studies employed both targeted and untargeted metabolomics techniques to explore metabolic changes in T. cruzi during stress conditions such as dietary deprivation and oxidative stress. These findings have shed light on the parasite's adaptation mechanisms in critical circumstances [5]. However, comprehensive protocols explaining the methodology for sample preparation, metabolite extraction, and subsequent analysis utilizing gas chromatography–mass spectrometry (GC–MS) remain scarce [6,7].
We describe a standard protocol for untargeted metabolomics analysis of T. cruzi epimastigotes grown in normal or varied culture conditions. Our method includes a thorough workflow from parasite cultivation and sample preparation to metabolite derivatization and GC–MS analysis. This methodology identifies metabolic alterations with excellent sensitivity and reproducibility, allowing researchers to investigate T. cruzi's metabolic flexibility, as well as other parasites, in response to environmental stresses [8]. Compared to earlier techniques, our protocol preserves parasite viability under stress circumstances, reduces sample variability, and provides a robust bioinformatics pipeline for data analysis [9].
Materials and reagents
Biological materials
1. Epimastigote forms of Trypanosoma cruzi DM28c strain (wild type) [10]
Reagents
1. Sodium chloride (Sigma-Aldrich, catalog number: S9888-1KG)
2. Potassium chloride (Sigma-Aldrich, catalog number: P3911-1KG)
3. Sodium phosphate dibasic (Sigma-Aldrich, catalog number: S9763-1KG)
4. Potassium phosphate (Sigma-Aldrich, catalog number: P0662-1KG)
5. BactoTM tryptose (BD DifcoTM, catalog number: 226920)
6. Liver infusion broth (BD DifcoTM, catalog number: 211713)
7. Calcium chloride (Sigma-Aldrich, catalog number: C1016-500G)
8. Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266-1KG)
9. Anhydrous sodium bicarbonate (Sigma-Aldrich, catalog number: 792519-1KG)
8. Hemin (Sigma-Aldrich, catalog number: H9039-1G)
9. Triethanolamine (Millipore, catalog number: 1.08379)
10. D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270-1KG)
11. Penicillin G (Sigma-Aldrich, catalog number: P3032-25MU)
12. Streptomycin sulfate, Streptomyces sp. (Sigma-Aldrich, catalog number: 5711)
13. Fetal bovine serum (FBS) (GibcoTM, catalog number: 210635K)
14. Glucose oxidase from Aspergillus niger (Sigma-Aldrich, catalog number: G7141-50UK)
15. PIPES (Sigma-Aldrich, catalog number: P8203-50G)
16. EGTA (Sigma-Aldrich, catalog number: E3889-500G)
17. Methanol, ≥ 99.8% (GC), HPLC grade, suitable for HPLC, LiChrosolv® (Supelco, catalog number: 1.06018)
18. O-methoxyamine hydrochloride [for GC derivatization, LiChropurTM, 97.5%–102.5% (AT)] (Supelco, catalog number: 89803)
19. Pyridine [puriss. p.a., ACS reagent, reag. Ph. Eur., ≥ 99.5% (GC)] (Sigma-Aldrich, catalog number: 33553)
20. N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) (for GC derivatization, LiChropurTM, 99.0%) (Supelco, catalog number: 15222-5ML-F) plus 1% trimethylchlorosilane (TMCS) (Pierce Chemical Co., Rockford, IL, USA)
21. Pentadecanoic acid (Sigma-Aldrich, catalog number: P6125-1G)
22. Heptane (hypergrade for LC-MS LiChrosolv®) (Sigma-Aldrich, catalog number: 1036541000)
23. Isopropanol [≥ 99.8% (GC), ACS reagent, reag. Ph. Eur., reag. ISO, EMSURE®] (Supelco, catalog number: 1096345005)
24. NaOH (Sigma-Aldrich, catalog number: 221465-1KG)
Solutions
1. Liver infusion tryptose (LIT) medium [11] (see Recipes)
2. Triatomine artificial urine medium (TAU) [12] (see Recipes)
3. Glucose oxidase solution (see Recipes)
4. Phosphate-buffered saline (PBS) [13] (see Recipes)
5. Hemin solution (see Recipes)
6. Methoximation solution (see Recipes)
7. Internal standard solution (IS) (see Recipes)
Recipes
1. Liver infusion tryptose (LIT) medium
Dissolve sodium chloride, potassium chloride, sodium phosphate, tryptose, and liver infusion broth in 800 mL of ultrapure water (Milli-Q), ensuring complete dissolution before adding hemin. Adjust to pH 7.2 and complete the solution volume to 1 L. Dispense the medium into 500 mL glass flasks and autoclave at 121 °C for 20 min. The medium can be stored at 4 °C for up to 3 months. Before use, supplement with glucose, fetal bovine serum, penicillin, and streptomycin. The supplemented medium should be stored at 4 °C for a maximum of 1 month. Before use, warm the medium to 28 °C, which is the optimal cultivation temperature for T. cruzi epimastigotes.
| Reagent | Weight | Volume |
|---|---|---|
| Sodium chloride | 2 g | - |
| Potassium chloride | 0.2 g | - |
| Sodium phosphate | 4 g | - |
| Tryptose | 2.5 g | - |
| Liver infusion broth | 2.5 g | - |
| Hemin 10 mg/mL (see Recipe 5) | - | 1 mL of 10 μg/mL stock |
| Glucose 20% (sterilized by filtration) | 0.2% | 10 mL of 0.2% |
| Fetal bovine serum (heat inactivated 30 min at 56 °C) | - | 100 mL |
| Penicillin (sterilized by filtration) | - | 1 mL of 59 mg/mL |
| Streptomycin (sterilized by filtration) | - | 1 mL 133 mg/mL |
| Total volume | - | 1,000 mL |
2. Triatomine artificial urine medium (TAU)
Dissolve calcium chloride, magnesium chloride, anhydrous sodium bicarbonate, sodium chloride, and potassium chloride in 800 mL of ultrapure water (Milli-Q). Adjust the pH to 6.2 using a 1 M sodium phosphate stock solution and bring the final volume to 1 L. Dispense the medium into 500 mL glass flasks and sterilize the solution using a 0.22 μm filter. The medium can be stored at 4 °C for up to 3 months. Before using it, warm it to 28 °C.
| Reagent | Weight | Volume |
|---|---|---|
| Calcium chloride | 0.294 g | - |
| Magnesium chloride | 0.406 g | - |
| Anhydrous sodium bicarbonate | 0.350 g | - |
| Sodium chloride | 11.1 g | - |
| Potassium chloride | 1.267 g | - |
| Total volume | - | 1,000 mL |
3. Glucose oxidase solution
To make the glucose oxidase (GOX) stock solution, dissolve PIPES in ultrapure water (Milli-Q) to a final concentration of 80 mM, add 1 mM EGTA and 1 mM MgCl2, and adjust the pH to 6.9 with NaOH in 100 mL, ensuring thorough dissolution. Then, dissolve glucose oxidase in 10 mL of this buffer to a final concentration of 6 U/mL, stirring gently to prevent enzyme denaturation. To avoid repeating the freeze-thaw cycle, divide the solution into small aliquots (500 μL) and store at -80 °C. To conduct experiments, dilute this stock solution 1:10 to a final working concentration of 0.6 U/mL in the culture medium.
| Reagent | Weight | Volume |
|---|---|---|
| PIPES | 2.42 g | - |
| EGTA | 38 mg | - |
| Magnesium chloride | 9 mg | - |
| 100 mL | ||
| Glucose oxidase | 60 U | - |
| Total | - | 10 mL |
4. Phosphate-buffered saline (PBS)
Dissolve sodium chloride, potassium chloride, sodium phosphate, and potassium phosphate in 1 L of ultrapure water (Milli-Q). Transfer the buffer to a 1 L glass flask and autoclave at 121 °C. Filter the solution through a 0.22 μm filter in a laminar airflow cabinet before aliquoting into 100 mL flasks. The sterile buffer should be kept at room temperature for a maximum of one month.
| Reagent | Weight | Volume |
|---|---|---|
| Sodium chloride | 8 g | - |
| Potassium chloride | 0.2 g | - |
| Sodium phosphate | 1.15 g | - |
| Potassium phosphate | 0.2 g | - |
| Total volume | - | 1,000 mL |
5. Hemin stock solution
Make a paste by combining 200 mg of hemin and 200 μL of triethanolamine. Dissolve the mixture in 20 mL of ultrapure water to 10 mg/mL. The solution can be kept at 4 °C for up to six months.
| Reagent | Weight | Volume |
|---|---|---|
| Hemin | 200 mg | - |
| Triethanolamine | - | 0.2 mL |
| Total volume | - | 20 mL |
6. Methoximation solution
Dilute O-methoxyamine hydrochloride in pyridine to 15 mg/mL and store at -20 °C.
| Reagent | Weight | Volume |
|---|---|---|
| O-methoxyamine hydrochloride | 0.045 g | - |
| Pyridine | - | 3 mL |
| Total volume | - | 3 mL |
7. Internal standard solution
Dilute pentadecanoic acid heptane at 1,000 ppm (1 g/L) to 10 ppm in heptane and store at -20 °C.
| Reagent | Weight | Volume |
|---|---|---|
| Pentadecanoic acid heptane | - | 100 μL |
| Heptane | - | 9.9 mL |
| Total volume | - | 10.0 mL |
Laboratory supplies
1. Cell culture flasks 75 cm2 (Biofil, catalog number: TCF001250)
2. Cell culture flasks 25 cm2 (Biofil, catalog number: TCF001050)
3. 15 mL graduated centrifuge tubes (Biofil, catalog number: CFT511150)
4. 50 mL graduated centrifuge tubes (Biofil, catalog number: CFT011500)
5. 5 mL serological pipettes (Biofil, catalog number: GSP010005)
6. 10 mL serological pipettes (Biofil, catalog number: GSP010010)
7. 25 mL serological pipettes (Biofil, catalog number: GSP010025)
8. 1–200 μL pipette tips (Corning, catalog number: 4711)
9. 100–1,000 μL pipette tips (Corning, catalog number: 4714)
11. Cell counting Neubauer chamber (Corning, catalog number: 480200)
12. P20 micropipette (Gilson, catalog number: F144056M)
13. P200 micropipette (Gilson, catalog number: F144058M)
14. P1000 micropipette (Gilson, catalog number: F144059M)
15. 1.5 mL microtube (Axygen, catalog number: MCT-150-A)
16. 1.5 mL cryotube (Corning, catalog number: 430659)
17. 1 L glass storage bottle (Pyrex, catalog number: CLS13951L)
18. 500 mL glass storage bottle (Pyrex, catalog number: CLS1395500)
19. 100 mL glass storage bottle (Pyrex, catalog number: CLS1395100)
20. Nitrile gloves (Supermax, catalog number: 011185)
21. 1 L vacuum filtration devices, pore 0.22 μm (Corning, catalog number: CLS430015-12EA)
Equipment
1. Laminar airflow cabinet (Trox, catalog number: 583/FLV)
2. Tabletop refrigerated centrifuge for 15 and 50 mL tubes (Thermo Scientific, model: Sorvall® LegendTM Mach 1.6R)
3. Microcentrifuge refrigerated for 1.5 and 2 mL microtubes (Thermo Scientific, model: Sorvall® LegendTM Micro 17R)
4. Vortex mixer (Daigger, model: Vortex Genie 2, catalog number: 22220A)
5. Ultrasonic bath (Synth, model: SSBuc 6l)
6. 4 °C refrigerator (Consul, catalog number: CDR46/127)
7. -20 °C freezer (Consul, catalog number: CDR46/127)
8. Ultra-low -80 °C freezer (Panasonic, catalog number: MDF U33V-PA, VIP Series)
9. Liquid nitrogen (N2) storage tank (Bridgepath® International Cryogenics Director D4000)
10. GC–MS chromatography coupled with a mass spectrometer quadrupole type (Shimadzu Co., model: QP2020NX)
11. SLBTM-5MS fused silica capillary column (30 m × 0.25 mm, 0.25 μm thickness, Supelco)
12. Helium gas 5.0 analytical grade (White Martins Ltda.)
13. Electronic balance (Bioprecisa, catalog number: FA 2104R)
14. SpeedVac concentrator (Eppendorf, model: Vacufuge®)
15. Fume hood (Braslab® Propylene Box)
16. 28 °C BOD incubator (Tecnal, model: TE-371)
17. 70 °C incubator (SureTemp® Dual Convection incubator, catalog number: Z742691-1EA)
18. Vacuum pump (Fanem® Diapump®, model 089/CA)
19. Serological pipette controller (HTL SwiftPet® Pro)
20. Racks (Prolab®)
Software and datasets
1. GCMS Solution (Shimadzu Co., version 4.52) (requires a license)
2. NIST17 MASS (Shimadzu Co., version 1.00.1) (requires a license)
3. GCMS Smart Metabolite (Shimadzu Co., version 3.01) (requires a license)
4. Excel (Microsoft Office) (requires a license)
5. https://www.genome.jp/kegg/ (free to use)
6. http://www.hmdb.ca (free to use)
7. http://www.metaboanalyst.ca/ (free to use)
8. Orange 3 (free to use)
Procedure
文章信息
稿件历史记录
提交日期: Mar 27, 2025
接收日期: May 14, 2025
在线发布日期: Jun 26, 2025
出版日期: Jul 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Silva, M. A., Izidoro, M., Bonifácio, B. S. and Schenkman, S. (2025). Untargeted Metabolomics of Epimastigote Forms of Trypanosoma cruzi. Bio-protocol 15(13): e5368. DOI: 10.21769/BioProtoc.5368.
分类
微生物学 > 微生物生理学 > 胁迫反应
系统生物学 > 代谢组学 > 全生物体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link