- EN - English
- CN - 中文
An Improved m6A-ELISA for Quantifying N6-methyladenosine in Poly(A)-purified mRNAs
用于Poly(A)纯化mRNA中N6-甲基腺苷定量的改良型m6A-ELISA方法
发布: 2025年06月20日第15卷第12期 DOI: 10.21769/BioProtoc.5359 浏览次数: 3006
评审: Joyce ChiuNavnita DuttaAnonymous reviewer(s)
Abstract
N6-methyladenosine (m6A) is an abundant internal mRNA modification with roles in regulating cellular and organismal physiology, including development, differentiation, and disease. The deposition of m6A is highly regulated, with various m6A levels across different environmental conditions, cellular states, and cell types. Available methods for measuring bulk m6A levels are often time-consuming, have low throughput, and/or require specialized instrumentation or data analyses. Here, we present a detailed protocol for measuring bulk m6A levels in purified poly(A) RNA samples with m6A-ELISA using a standard-based approach. Critical steps of the protocol are highlighted and optimized, including poly(A) RNA quality controls and antibody specificity testing. The protocol is fast, scalable, adaptable, and cost-effective. It does not require specialized instrumentation, training, or skills in data analysis. We have successfully tested this protocol on mRNAs isolated from budding yeast and mouse cell lines.
Key features
• N6-methyladenosine quantification, including mRNA isolation, can be achieved in two days.
• Describes a robust method for poly(A) RNA isolation from total RNA, minimizing non-poly(A) RNA contamination.
• Based on Ensinck et al. [1], optimizing poly(A) RNA selection from yeast and mammalian cells and reducing the amount of mammalian mRNA for the assay.
Keywords: ELISA (ELISA)Graphical overview
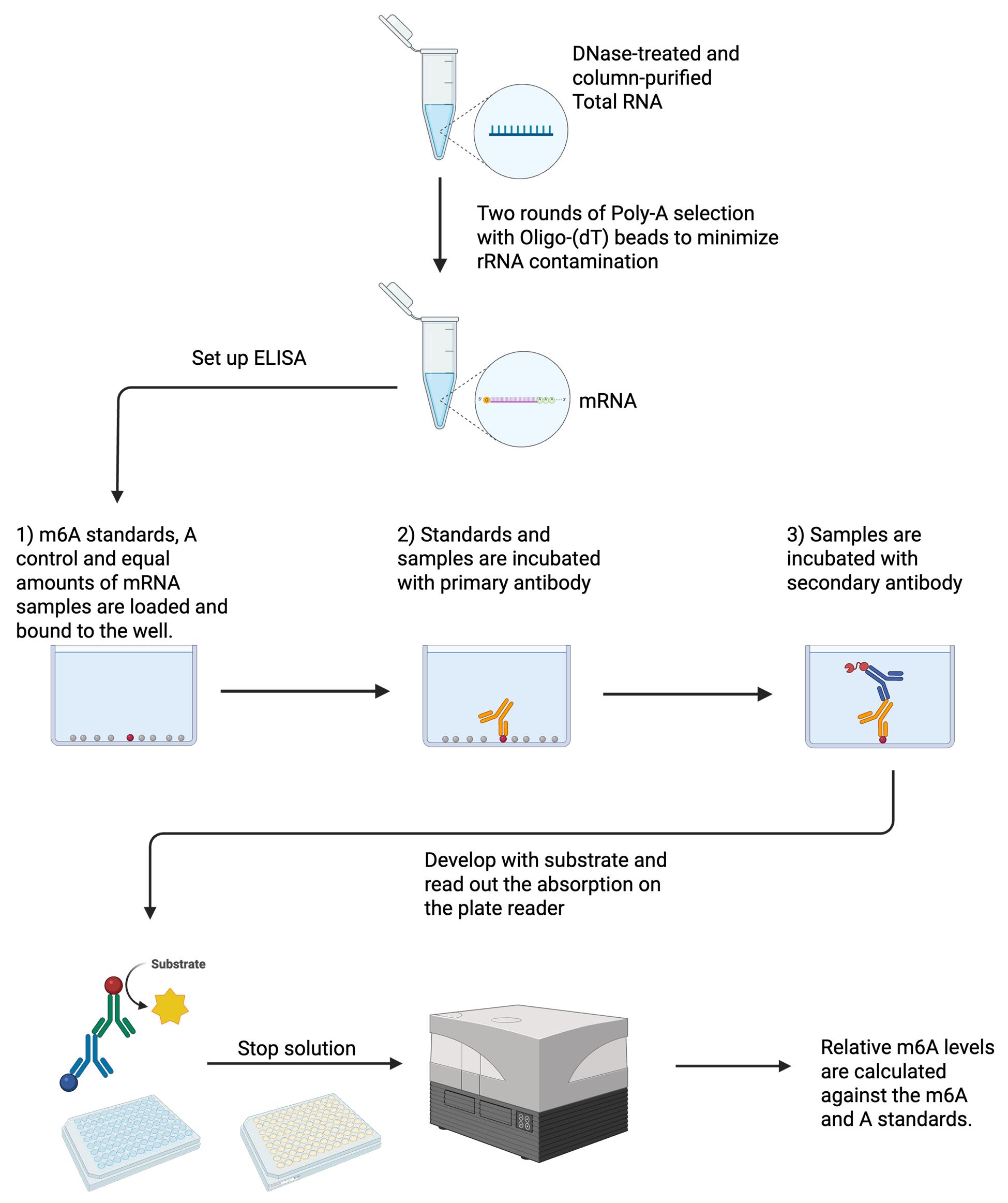
Total RNA is extracted, DNase-treated, and column-purified. Two rounds of poly(A) selection are performed to minimize rRNA contamination. Equal amounts of mRNA between samples are bound to wells on a 96-well plate. In vitro transcribed A and m6A are also bound as standards. Samples are incubated with the primary antibody and, subsequently, with the secondary antibody. Samples are developed with a substrate, stop solution is added, and absorption is read out on a plate reader. Relative m6A levels are calculated against the m6A and A standards. Created in BioRender. Chan, W. (2025) https://BioRender.com/e69h636
Background
Epitranscriptomics, post-transcriptional chemical RNA base modifications, have emerged as key regulators of gene expression. Among RNA modifications, N6-methyladenosine (m6A) is a prevalent internal mRNA modification that is deposited by a multisubunit methyltransferase complex (m6A writers) on the RRACH motif sequence in mRNAs [2–4]. The m6A mark is then recognized and bound by various proteins, including a class of proteins containing the YTH (YT521-B homology) domain, also known as m6A readers, and directs m6A-modified transcripts for subsequent fates such as decay, translation, and localization [5–7]. Another group of mRNA-modifying enzymes known as m6A erasers can remove the m6A modification [8]. Together, they make up the m6A machinery, which dynamically regulates m6A levels. As a result, m6A levels may vary substantially across different environmental conditions, cell types, and species. For example, in Saccharomyces cerevisiae, m6A is specifically deposited during early meiosis but is absent in other cellular states. The dynamic regulation of m6A is critical for maintaining stem cells and regulating neuronal cell differentiation, development, stress response, and the immune system [9,10]. Misregulation of m6A is implicated in pathology, including that of inflammatory and immune deregulatory diseases and cancer [11].
Various methods for quantifying m6A levels are available, ranging from total or global m6A levels to single-nucleotide m6A quantification. Several sequencing-based approaches exist, including antibody-based approaches (such as m6A-seq [12] and miCLIP [13]), and antibody-free methods (such as GLORI-seq [14], MATZER-seq [15], eTAM-seq [16], and Nanopore [17]). Sequencing methods can be time-consuming, costly, and require specialized data analysis. Global m6A detection methods, on the other hand, do not require sequencing or extensive data analysis, with the most accurate being mass spectrometry (MS), where m6A nucleosides from digested purified mRNA are quantified by LC/MS [18,19]. Thin-layer chromatography [20] and m6A dot blot [21] have also been used for total m6A detection and relative quantification. However, drawbacks of the aforementioned methods include MS requiring specialist training and equipment, whereas thin-layer chromatography and m6A dot blot are typically low throughput and low in sensitivity.
We have previously shown that m6A-ELISA is a simple quantitative method for detecting global m6A levels [1]. m6A-ELISA is a cost-effective and fast approach that does not require specialist training or instruments [1]. Here, we describe a step-by-step protocol of m6A-ELISA, including a detailed description of the procedures for poly(A) selection that yields high-quality mRNA and m6A antibody validation. We optimized the poly(A) mRNA purification step and enhanced the sensitivity for m6A detection.
Materials and reagents
Biological materials
1. DNase-treated and column-purified total RNA from samples to be measured
2. DNase-treated and column-purified total RNA from ime4Δ yeast strain (Horizon, catalog number: YSC62750201935429), for antibody blocking. If working with a strain other than BY4743, we recommend generating an ime4 IME4 deletion strain following the PCR-mediated gene disruption protocol described in [22], Gene 1995; Jan 151(1):113–117. https://doi.org/10.1016/0378-1119(95)00144-U. For culture conditions, please refer to [3] eLife 2023; NaN:RP87860. https://doi.org/10.7554/eLife.87860
Reagents
1. Nuclease-free (NF) water (Ambion, catalog number: AM9932)
2. Lithium chloride, 8 M solution (Sigma-Aldrich, catalog number: L7026)
3. Lithium dodecyl sulfate (LiDS) (Thermo Fisher Scientific, catalog number: 413301000)
4. Tris-HCl, pH 7.4, 1 M (Sigma-Aldrich, catalog number: T21941)
5. EDTA, 0.5 M solution, pH 8.0 (Invitrogen, catalog number: 2624961)
6. 10× PBS (Thermo Fisher Scientific, catalog number: AM9624)
7. Polysorbate 20, e.g., Tween® 20 (Sigma-Aldrich, catalog number: P9416)
8. Oligo-d(T)25 magnetic beads (New England BioLabs, catalog number: S1419S)
9. Qubit® RNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q32855)
10. DNA binding microplate solution (Abcam, catalog number: ab156917)
11. N6-methyladenosine (m6A) rabbit mAb (ABclonal, catalog number: A19841)
12. Goat anti-rabbit IgG H&L (HRP) (Abcam, catalog number: ab205718)
13. TMB ELISA substrate (fast kinetic rate) (Abcam, catalog number: ab171524)
14. 450 nm Stop solution for TMB substrate (Abcam, catalog number: ab171529)
15. MEGAscript T7 Transcription kit, 25 reactions (ThermoFisher Scientific, catalog number: AM1333)
16. N6-Methyl-ATP (Jena Bioscience, catalog number: NU-1101L)
17. TURBOTM DNase (ThermoFisher Scientific, catalog number: AM2238)
18. Macherey-NagelTM NucleoSpinTM RNA Clean-up, Mini kit for RNA clean up and concentration (Macherey-Nagel, catalog number: 740948.250)
Solutions
1. Binding buffer (see Recipes)
2. Wash buffer A (see Recipes)
3. Wash buffer B (see Recipes)
4. Primary antibody solution blocked with total m6A-free RNA (DNase-treated and column-purified) ime4Δ (see Recipes)
5. Secondary antibody solution (see Recipes)
Recipes
Note: The total volume needed will depend on the number of samples. We recommend all buffers be made up fresh for short-term use and filter-sterilized. The tables below describe the recipes for 50 mL; volumes can be scaled up or down as needed.
1. Binding buffer (50 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Tris-HCl, pH 7.4, 1 M | 20 mM | 1 mL |
| Lithium chloride, 8 M | 500 mM | 3.125 mL |
| Lithium dodecyl sulfate (10% w/v) | 0.5% | 2.5 mL |
| EDTA, 0.5 M | 2 mM | 200 μL |
| NF water | 43.175 mL |
Storage: Short-term (i.e., up to 1 week) at 4 °C.
Caution: The LiDS may precipitate at 4 °C; this can be resolved by bringing the solution to room temperature.
2. Wash buffer A (50 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Tris-HCl, pH 7.4, 1 M | 20 mM | 1 mL |
| Lithium chloride, 8 M | 500 mM | 3.125 mL |
| Lithium dodecyl sulfate (10% w/v) | 0.2% | 1 mL |
| EDTA, 0.5 M | 2 mM | 200 μL |
| NF water | 44.675 mL |
Storage: Short-term (i.e., up to 1 week) at 4 °C.
Caution: The LiDS may precipitate at 4 °C; this can be resolved by bringing the solution to room temperature.
3. Wash buffer B (50 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| Tris-HCl, pH 7.4, 1 M | 20 mM | 1 mL |
| Lithium chloride, 8 M | 500 mM | 3.125 mL |
| EDTA, 0.5 M | 2 mM | 200 μL |
| NF water | 45.675 mL |
Storage: Short-term (i.e., up to 1 week) at 4 °C.
4. Primary antibody solution blocked with total m6A-free RNA (DNase-treated and column-purified) ime4Δ (10 mL)
| Reagent | Final concentration | Quantity |
|---|---|---|
| 1× PBS 0.1% Tween 20 | 1× | 10 mL |
| N6-methyladenosine (m6A) rabbit mAb | 1:10,000 | 1 μL |
| Total m6A-free RNA (DNase-treated and column clean-up from ime4Δ yeast strain) | 500 ng·mL-1 | 5 μg |
Storage: Short-term (i.e., during setup of the ELISA) at 4 °C. We do not recommend reusing the primary antibody.
5. Secondary antibody solution (10 mL)
| Reagent | Final concentration | Volume |
|---|---|---|
| 1× PBS 0.1% Tween 20 | 1× | 10 mL |
| Goat anti-rabbit IgG H&L (HRP) | 1:5,000 | 2 μL |
Storage: Short-term (i.e., during setup of the ELISA) at 4 °C. We do not recommend reusing the secondary antibody.
Laboratory supplies
1. Low-retention 300 μL pipette tips (Elkay, catalog number: AER-4REF-S96)
2. Low-retention 1,000 μL pipette tips (Elkay, catalog number: AER-STE1-A18)
3. 10 μL filter pipette tips (Starlab, catalog number: S1121-3810-C)
4. 200 μL filter pipette tips (Starlab, catalog number: S1123-8810-C)
5. 1,000 μL filter pipette tips (Starlab, catalog number: S1126-7710-C)
6. 1.5 mL DNA LoBind Eppendorf tubes (snap cap) (Eppendorf, catalog number: 0030108051)
7. Qubit® assay tubes (Thermo Fisher Scientific, catalog number: Q32856)
8. 96-well microplate pack (Abcam, catalog number: ab210903)
9. Milipore® Stericup® quick release vacuum filtration system (Merck, catalog number S2GPU0SRE)
10. Adhesive PCR plate seals (Thermo Fisher Scientific, catalog number: AB0558)
Equipment
1. Eppendorf Thermomixer, 24 × 1.5 mL microcentrifuge tubes (or equivalent alternative)
2. DynaMag-2 magnetic rack (Thermo Fisher Scientific, catalog number: 12321D) (or equivalent alternative)
3. Nanodrop One spectrophotometer (Thermo Fisher Scientific, catalog number: ND-ONE-W) (or equivalent alternative)
4. Rotator wheel
5. Multi-channel pipette 30–300 μL (Eppendorf, catalog number: 3125000052) (or equivalent alternative)
6. Qubit 4 fluorometer (Thermo Fisher Scientific, catalog number: Q33238)
7. 450 nm plate reader, e.g., Tecan Spark® microplate reader (or equivalent alternative)
Software and datasets
1. 450 nm plate reader software, e.g., Tecan Spark Control version 3.0 (or equivalent alternative)
2. Excel Spreadsheet (Microsoft)
3. GraphPad Prism (GraphPad Software) (optional)
Procedure
文章信息
稿件历史记录
提交日期: Feb 13, 2025
接收日期: May 19, 2025
在线发布日期: Jun 9, 2025
出版日期: Jun 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Chan, W. Y., Albihlal, W. S. and Van Werven, F. J. (2025). An Improved m6A-ELISA for Quantifying N6-methyladenosine in Poly(A)-purified mRNAs. Bio-protocol 15(12): e5359. DOI: 10.21769/BioProtoc.5359.
分类
分子生物学 > RNA > RNA 修饰
系统生物学 > 表观基因组学
生物化学 > RNA > RNA-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













