- EN - English
- CN - 中文
Isolation and Characterization of Cervical Cancer-Associated Mesenchymal Stem Cells From Primary Tumors Using Explant Culture
基于外植体培养法从宫颈癌原发瘤中分离并鉴定癌相关间充质干细胞
(*contributed equally to this work) 发布: 2025年06月20日第15卷第12期 DOI: 10.21769/BioProtoc.5358 浏览次数: 2745
评审: Elena A. OstrakhovitchMithun Santra
Abstract
Cancer-associated mesenchymal stem cells (Ca-MSCs), an integral part of the tumor microenvironment, play a major role in modulating tumor progression; they have been reported to progress as well as inhibit various cancers, including cervical cancer. To understand the exact role of Ca-MSCs in tumor modulation, it is necessary to have an optimized protocol for Ca-MSCs isolation. This work demonstrates the isolation and expansion of a primary culture of cervical cancer–associated MSCs (CCa-MSCs) from the biopsy sample of cervical cancer patients using the explant culture technique. The isolated cells were characterized according to International Society for Cellular Therapy (ISCT) guidelines. Morphological analysis revealed that cells were adherent to the plastic surface and possessed spindle-shaped morphology. Flow cytometry analysis of the cells showed high expression (~98%) for MSC-specific cell surface markers (CD90, CD73, and CD105), negative expression (<0.5%) for endothelial cell marker (CD34) and hematopoietic cell marker (CD45), and negligible expression for HLA-DR, as recommended by ISCT. Further, trilineage differentiation potential analysis of the cells showed their osteogenic and chondrogenic potential and adipogenic differentiation. This standardized protocol will assist in the cultivation of CCa-MSCs and the study of their interactions with tumor cells and other components of the tumor microenvironment. This protocol may be utilized in the establishment of Ca-MSCs from other types of cancers as well.
Key features
• Isolation and expansion of cervical cancer–associated mesenchymal stem cells (CCa-MSCs) from patient biopsy sample.
• Characterization of isolated CCa-MSCs for the presence of MSC-specific cell surface markers and trilineage differentiation potential.
• CCa-MSCs can be employed to study the interactions with the tumor cells in the tumor microenvironment.
Keywords: Mesenchymal stem cells (间充质干细胞)Graphical overview
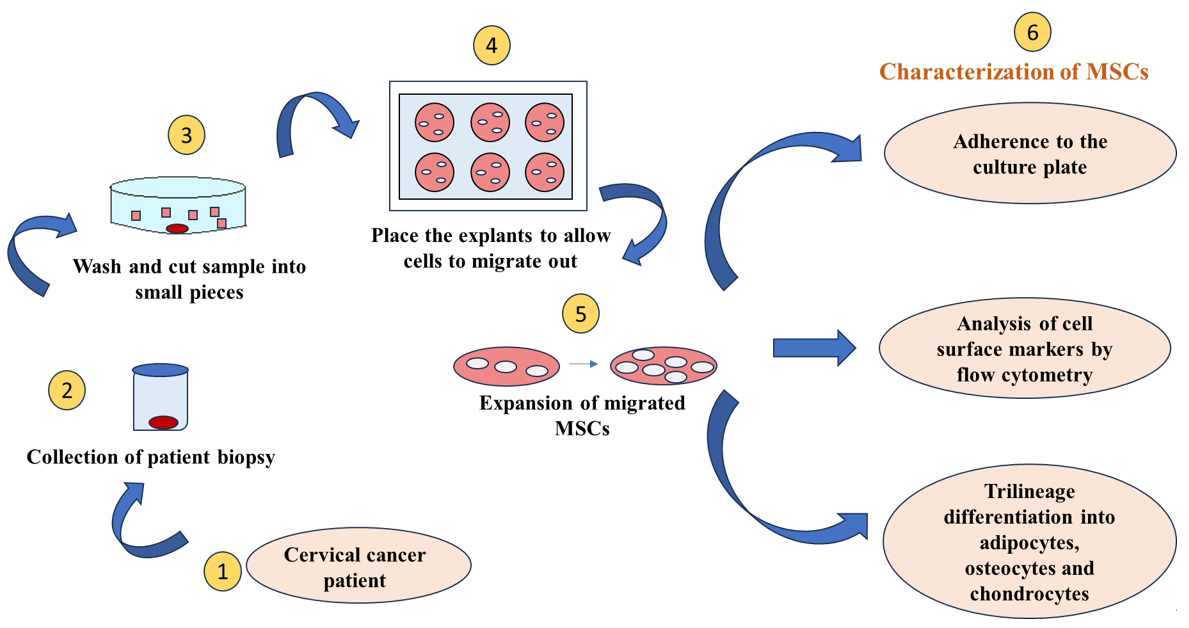
Background
A tumor is a micro-niche in which the cancer cells interact with their neighboring non-tumorigenic host cells. These intercellular communications are critical to the pathophysiology of this disease, and hence, they have gained much attention among researchers over the past couple of decades. The tumor niche, popularly termed tumor microenvironment (TME), comprises endothelial cells, fibroblasts, immune cells, and mesenchymal stem cells (MSCs), among others. It also consists of extracellular components like extracellular matrix, cytokines, or growth factors [1,2]. All these components are present in the vicinity of cancer cells and are further supported by a dense vascular network. This complex TME can modulate tumor initiation, progression, and metastasis and even alter the response of these cancer cells to therapy [3].
Currently, there is limited understanding of the interactions that occur within the TME. One such interaction that modulates tumor growth is one with MSCs, which, over the years, have been reported to act as a double-edged sword for participating both in tumor progression and inhibition [4]. MSCs are spindle-shaped cells that are present in almost all tissues of the body; with their high migratory abilities, they move to the site of the tumor. Numerous studies have been conducted to gain insight into the role of these MSCs in tumor progression, mainly using bone marrow–derived MSCs or adipose-derived MSCs (ADSCs) [5,6]. However, recent studies suggest that cancer-associated MSCs (Ca-MSCs) possess stronger pro-tumorigenic, immunosuppressive, and angiogenic properties than parental MSCs [1,7,8]. Studies suggest that MSCs migrated toward tumor sites differentiate into tumor-associated phenotypes to form Ca-MSCs. Even within the TME, Ca-MSCs have been reported to be educated by tumor cells, and they, in turn, secrete growth factors and chemokines that support tumor growth [9]. This indicates that MSCs isolated from tumor lesions truly represent the characteristics of TME rather than MSCs isolated from tumor-free tissues [10,11]. While the role of Ca-MSCs has been investigated in ovarian, gastric, hepatocellular, breast, and lung cancers, their role in cervical cancer remains largely unexplored [12–15].
Resistance to chemotherapy and radiotherapy remains a significant challenge, leading to tumor relapse in cervical cancer patients. This resistance is largely attributed to cancer stem cells within the TME, a small but highly potent subpopulation with strong self-renewal, differentiation, and tumorigenic potential [16]. Research aims to understand their resistance mechanisms and develop targeted therapies, necessitating an optimized protocol for isolating pure cancer stem cells. While Ca-MSCs have been isolated from various cancers [9], their characterization in cervical cancer remains limited. To address this, we focused on isolating and characterizing cervical cancer–associated mesenchymal stem cells (CCa-MSCs) using an optimized explant culture method.
Previously, Garcia et al. reported the isolation of CCa-MSCs using enzymatic digestion alone [17]. However, this method may compromise cell viability due to cellular damage. Research suggests that explant culture yields a more proliferative, homogenous cell population compared to enzymatic digestion [18]. Accordingly, in the present study, we have successfully demonstrated the isolation and characterization of CCa-MSCs by the explant culture method which generates a homogenous cell population free from contamination of other cell types [17–20].
Materials and reagents
Biological materials
1. Biopsy samples were collected from three treatment-naive cervical cancer patients diagnosed with squamous cell carcinoma or adenocarcinoma (Table 1). The sample was collected by the consulting gynecologist in the Department of Obstetrics and Gynecology, PGIMER Chandigarh, after obtaining proper informed consent and approval by the Institute Ethics Committee.
Table 1. Clinical details of the cervical cancer patients
| Patient ID | Age of the patient | Cervical cancer subtype |
|---|---|---|
| P1 | 54 | Adenocarcinoma |
| P2 | 65 | Moderately differentiated squamous cell carcinoma |
| P3 | 49 | Moderately differentiated squamous cell carcinoma |
Reagents
1. Alizarin red S stain (Sigma-Aldrich, catalog number: TMS-008)
2. Antibiotic antimycotic cocktail (Himedia, catalog number: A002-5X50ML)
3. Ascorbic acid (Himedia, catalog number: TC094-25G)
4. Dexamethasone 21-phosphate sodium (Sigma-Aldrich, catalog number: D1159-100MG)
5. DMEM low glucose (Gibco, catalog number: 11885-084)
6. Dulbecco’s phosphate-buffered saline (PBS) (Himedia, catalog number: TS1006-5L)
7. EZ Stain Chondrocyte Staining kit (Alcian blue stain) (Himedia, catalog number: CCK029)
8. Fetal bovine serum (FBS) (Gibco, catalog number: 16000044)
9. Human TGF-β3 (Peprotech, catalog number: 100-36E-10U)
10. Insulin-Transferrin-Selenous acid premix (Sigma-Aldrich, catalog number: I3146)
11. Monopotassium phosphate (Sigma-Aldrich, catalog number: 15655-100G)
12. Oil red O stain (Sigma-Aldrich, catalog number: 01391-250Ml)
13. Sodium pyruvate solution (Sigma-Aldrich, catalog number: S8636-100ML)
14. StemPro Adipogenesis Differentiation kit (Thermo Fisher Scientific, catalog number: A1007001)
15. TrypLE Express enzyme (Gibco, catalog number: 12604021)
16. β-glycerophosphate disodium salt hydrate (Sigma-Aldrich, catalog number: G9422-50G)
17. 10% neutral buffered formalin solution (Sigma-Aldrich, catalog number: HT501128-4L)
Antibodies
1. PE Mouse anti-human CD105 antibody (1:50) (BD-Pharmingen, catalog number: 560839)
2. FITC anti-human CD34 antibody (1:50) (BioLegend, catalog number: 343604)
3. FITC anti-human CD45 antibody (1:50) (BioLegend, catalog number: 304006)
4. FITC anti-human CD73 antibody (1:50) (BioLegend, catalog number: 344016)
5. FITC anti-human CD90 antibody (1:50) (BioLegend, catalog number: 328107)
6. PE anti-human HLA-DRHLA-DR (1:50) (BioLegend, catalog number: 307065)
Solutions
1. CCa-MSCs culture media (see Recipes)
2. PBS (see Recipes)
3. Chondrogenic differentiation media (see Recipes)
4. Osteogenic differentiation media (see Recipes)
5. Osteocyte staining solution (Alizarin Red S stain) (see Recipes)
6. Adipocyte staining solution (Oil Red O stain) (see Recipes)
Recipes
1. CCa-MSCs culture media
Supplement DMEM (1 g/L) with 10% FBS and 1× antibiotic antimycotic solution (100 U penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin B).
2. PBS
Dissolve 9.6 g of PBS in sterile water and autoclave. Add 1× antibiotic antimycotic solution (100 U penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin B). Filter the PBS solution.
3. Chondrogenic differentiation media
| Reagent | Stock concentration | Final concentration | Volume |
|---|---|---|---|
| Ascorbic acid | 200 M | 0.2 M | 50 μL |
| Sodium pyruvate | 100 mM | 1 mM | 500 μL |
| Human TGF-β3 | 250 mM | 10 μM | 2 μL |
| Dexamethasone 21-phosphate sodium | 10 M | 1 mM | 5 μL |
| Insulin-Transferrin-Selenous acid premix | 1,000× | 1× | 50 μL |
| DMEM low glucose | n/a | n/a | 50 mL |
| Antibiotic-antimycotic solution | 100× | 1× | 500 μL |
| Total | n/a | 50 mL |
Note: Filter the differentiation media using a 0.22 μm syringe filter after mixing all the components. Prepare the stock solutions in sterile water. Individual components can be stored at -20 °C. Check for any precipitates after freeze thaw.
4. Osteogenic differentiation media
| Reagent | Stock concentration | Final concentration | Volume |
|---|---|---|---|
| β-glycerophosphate disodium salt hydrate | 2.5 M | 5 mM | 100 μL |
| Monopotassium phosphate | 0.5 M | 1.8 mM | 100 μL |
| Dexamethasone 21-phosphate sodium | 1 mM | 0.01 mM | 0.25 μL |
| DMEM low glucose | n/a | n/a | 50 mL |
| Antibiotic-antimycotic solution | 100× | 1× | 500 μL |
| Total | n/a | 50 mL |
Note: Filter the differentiation media using a 0.22 μm syringe filter after mixing all the components. Prepare the stock solutions in sterile water. Individual components can be stored at -20 °C. Check for any precipitates after freeze thaw.
5. Osteocyte staining solution (Alizarin Red S stain)
Note: Adjust the pH of the staining solution to 4.1–4.3 with 0.1% ammonium hydroxide or 0.1% HCl. Make up the total volume to 100 mL after adjusting the pH of the solution. Filter the solution through Whatman filter paper to remove any undissolved components.
| Reagent | Final concentration | Quantity |
|---|---|---|
| Alizarin Red S | n/a | 2 g |
| dH2O | n/a | 100 mL |
| Total | n/a | 100 mL |
6. Adipocyte staining solution (Oil Red O stain)
Note: Filter the solution through Whatman filter paper to remove any undissolved components.
| Reagent | Final concentration | Volume |
|---|---|---|
| Oil Red O stain | n/a | 30 mL |
| dH2O | n/a | 60 mL |
| Total | n/a | 90 mL |
Laboratory supplies
1. 6-well cell culture plate (Corning, catalog number: 3516)
2. T-25 flasks (Corning, catalog number: 430639)
3. T-75 flasks (Corning, catalog number: 430641U)
4. Petri dish (Tarson, catalog number: 460090-90MM)
5. 0.22 μm Whatman filter paper (Merck, catalog number: WHA1001325)
Equipment
1. EVOS LED microscope (Invitrogen)
2. FACS Canto flow cytometer (BD Biosciences)
3. Inverted microscope (Leica)
4. Centrifuge (ifuge L400P, model: LC01-3020 EU/AUS/IND)
5. Biosafety cabinet (Nuaire, model: NU-437-4009)
6. Surgical tools (scalpel and forceps)
7. CO2 incubator (Nuaire)
8. Hemocytometer (depth 0.1 mm) (Perfect Improved Neubauer)
9. -20 °C freezer (Vest Frost Solutions)
10. -80 °C freezer (Thermo Scientific)
11. Refrigerator (Samsung, model: RT28M3022S8/HL/2018)
12. 0.5–10 μL single channel variable pipette (Eppendorf, model: Research® plus, catalog number: 3125000028)
13. 10–100 μL single channel variable pipette (Eppendorf, model: Research® plus, catalog number: 3123000047)
14. 100–1,000 μL single channel variable pipette (Eppendorf, model: Research® plus, catalog number: 3123000063)
15. Autoclave (Equitron)
16. Mr. FrostyTM freezing container (Thermo Scientific, catalog number: 5100-0001)
17. Weighing balance (Wensar)
Software and datasets
1. BD FACSDiva 8.0.3
Procedure
文章信息
稿件历史记录
提交日期: Dec 15, 2024
接收日期: May 18, 2025
在线发布日期: Jun 12, 2025
出版日期: Jun 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Singla, S., Khurana, S., Bagga, R., Srinivasan, R. and Bhattacharyya, S. (2025). Isolation and Characterization of Cervical Cancer-Associated Mesenchymal Stem Cells From Primary Tumors Using Explant Culture. Bio-protocol 15(12): e5358. DOI: 10.21769/BioProtoc.5358.
分类
癌症生物学 > 癌症干细胞 > 肿瘤微环境
细胞生物学 > 细胞分离和培养 > 单层培养
干细胞 > 成体干细胞 > 癌症干细胞
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link