- EN - English
- CN - 中文
Using a Live Analysis System to Study Amyloplast Replication in Arabidopsis Ovule Integuments
利用活细胞成像系统研究拟南芥珠被中淀粉体的复制
发布: 2025年06月05日第15卷第11期 DOI: 10.21769/BioProtoc.5333 浏览次数: 2533
评审: Ansul LokdarshiSriema L. WalawageWenyang Li
Abstract
Amyloplasts, non-photosynthetic plastids specialized for starch synthesis and storage, proliferate in storage tissue cells of plants. To date, studies of amyloplast replication in roots and the ovule nucelli from various plant species have been performed using electron and fluorescence microscopy. However, a complete understanding of amyloplast replication remains unclear due to the absence of experimental systems capable of tracking their morphology and behavior in living cells. Recently, we demonstrated that Arabidopsis ovule integument could provide a platform for live-cell imaging of amyloplast replication. This system enables precise analysis of amyloplast number and shape, including the behavior of stroma-filled tubules (stromules), during proplastid-to-amyloplast development in post-mitotic cells. Here, we provide technical guidelines for observing and quantifying amyloplasts using conventional fluorescence microscopy in wild-type and several plastid-division mutants of Arabidopsis.
Key features
• Novel approach for investigating amyloplast differentiation and replication in plant cells.
• Detection of stroma-labeled amyloplasts in whole-mount ovules using conventional fluorescence microscopy.
• Facilitates quantitative and comparative analysis of amyloplast proliferation using various Arabidopsis resources.
• Enables high-resolution analysis of changing amyloplast and stromule morphologies in living cells.
Keywords: Amyloplast (淀粉体)Graphical overview
Pistils or siliques are dissected and subjected to conventional fluorescence microscopy for amyloplast observation and quantification in living tissue
Background
Plastids are double membrane–bound organelles found in the cells of seed plants that exhibit unique features of functional and morphological differentiation. These include chloroplasts in leaves, chromoplasts in flowers and fruits, and leucoplasts in general non-photosynthetic tissues [1]. Amyloplasts are non-photosynthetic plastids specialized for starch synthesis and storage. Amyloplasts develop from undifferentiated proplastids in meristematic tissues and are distributed in storage tissues such as roots, stems, and seeds, as well as in gravity-sensing tissues (for example, the stem endodermis and root cap columella) [2]. As starch is an important source of nutrients in human diets, the differentiation and proliferation of amyloplasts have long been an important area of crop biology research.
Plastids possess their DNA and typically replicate by binary fission [3,4]. In leaf mesophyll cells of the model plant Arabidopsis thaliana, chloroplast division is initiated by the formation of the stromal FtsZ ring and involves approximately 20 nuclear-encoded gene products located in the stroma, inner-envelope membrane, outer-envelope membrane, or cytoplasm [5,6]. In contrast, the mechanisms governing amyloplast proliferation remain largely unknown, partly due to the lack of an experimental system to track their number and behavior in specific cells.
We recently proposed that the Arabidopsis ovule integument could provide a platform for live-cell imaging of amyloplast replication and morphology, including the dynamics of stroma-filled tubules (stromules) [7]. This method may open a way to analyzing the function and regulation of plastid-division proteins during amyloplast proliferation in non-photosynthetic cells. In this protocol, we provide technical tips for conventional fluorescence microscopy of integument amyloplasts using wild-type (WT) and plastid-division mutants of Arabidopsis.
Materials and reagents
Biological materials
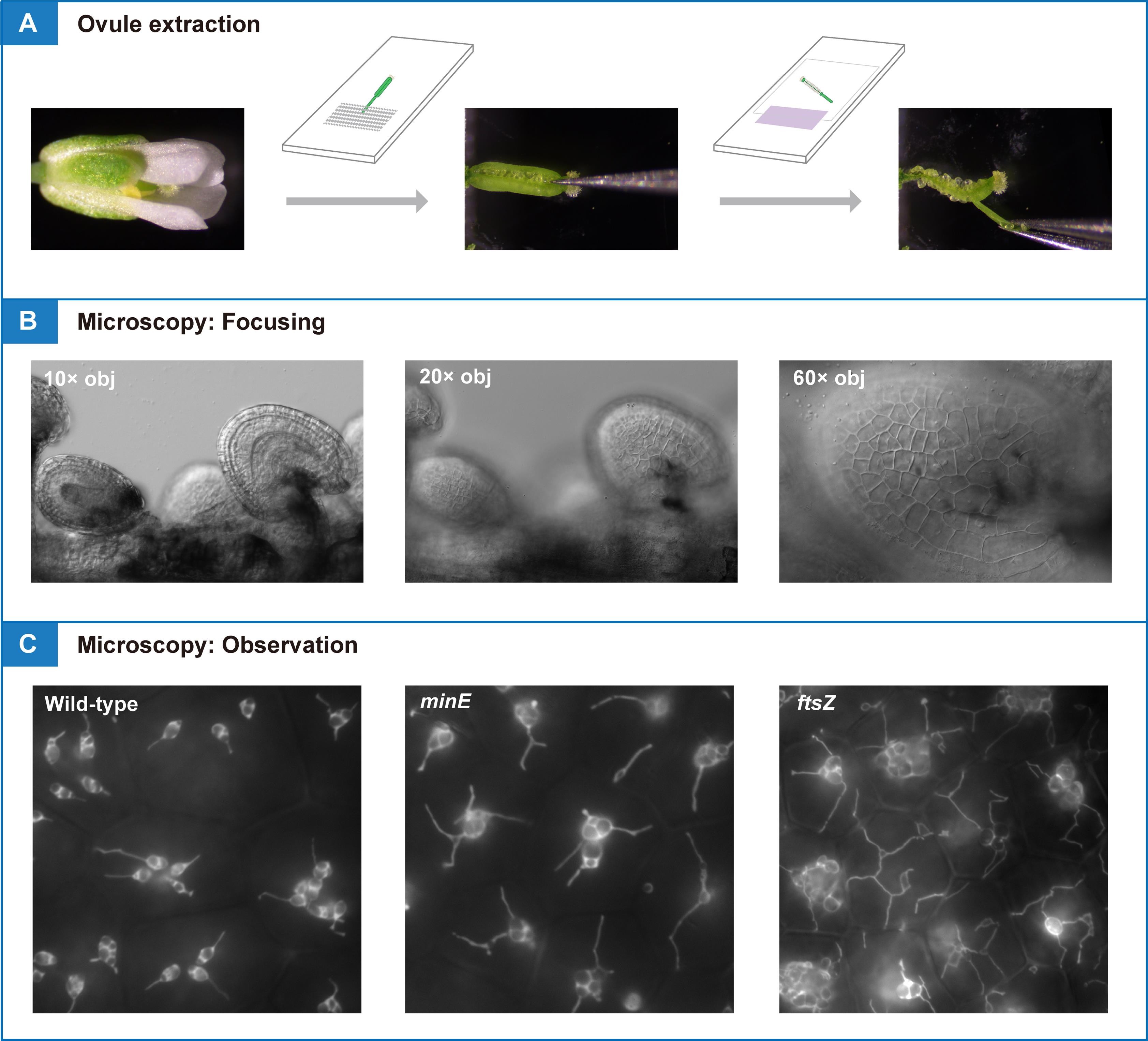
1. Seeds of transgenic Arabidopsis lines with stable expression of stroma-targeted fluorescent proteins (see General note 1). This protocol presents data from transgenic lines in the Col background [8] and minE [9], and ftsZ triple [10] (hereafter, ftsZ) mutants, in which a transit peptide (the N-terminal 90 aa of AtFtsZ1-1 [8])-fused YFP is driven by the constitutive CaMV35S promoter (see Figure 1)

Figure 1. Reproductive stages of Arabidopsis suitable for analysis of amyloplast replication. (A) Primary inflorescence. (B) Flower stages 14–17 [11] used for ovule extraction. (C) Expression pattern of CaMV35S promoter-driven stroma-targeted YFP in a transgenic Arabidopsis line. Images of brightfield (BF) and yellow fluorescent protein (YFP) signals are shown. Scale bar, 5 mm.
Reagents
1. Murashige and Skoog plant salt mixture (Wako Pure Chemical Industries, catalog number: 392-00591)
2. Sucrose (Wako Pure Chemical Industries, catalog number: 196-00015)
3. MES (Nacalai Tesque, catalog number: 21623-26)
4. Phyto agar (Duchefa Biochemie, catalog number: P1003.1000)
5. Jiffy-7 peat pellets, 44 × 42 mm (Jiffy International)
Laboratory supplies
1. Slide glass (Matsunami Glass, catalog number: S-2215)
2. Cover glass, 24 × 32 No. 1, thickness 0.13–0.17 mm (Matsunami Glass, catalog number: C024321)
3. Micropore surgical tape (3M, catalog number: 1530SP-0)
4. Paper tape (As One, catalog number: 6-691-01)
Equipment
1. Stereomicroscope with 0.63× objective lens (Leica Microsystems, model: MZ10 F)
2. CCD camera (Olympus, model: DP26) with 0.63× adaptor, attached to Leica MZ10 F
3. Inverted fluorescence microscope with 10×, 20×, and 60× objective lenses (Olympus, model: IX71)
4. CMOS camera (Hamamatsu Photonics, model: ORCA-flash2.8) with a 1.0× adaptor, attached to Olympus IX71
5. Tweezers 1 (Fontax, No. 5 Taxal)
6. Tweezers 2 (Dumont, No. DU-55 Dumoxel)
Software and datasets
1. Imaging software CellSens (Olympus, controller for DP26 camera)
2. Imaging software HCImage (Hamamatsu Photonics, controller for ORCA-flash2.8 camera)
Procedure
文章信息
稿件历史记录
提交日期: Jan 29, 2025
接收日期: May 6, 2025
在线发布日期: May 22, 2025
出版日期: Jun 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Fujiwara, M. T., Arakawa, R., Abe, T. and Itoh, R. D. (2025). Using a Live Analysis System to Study Amyloplast Replication in Arabidopsis Ovule Integuments. Bio-protocol 15(11): e5333. DOI: 10.21769/BioProtoc.5333.
- Fujiwara, M. T., Yoshioka, Y., Kazama, Y., Hirano, T., Niwa, Y., Moriyama, T., Sato, N., Abe, T., Yoshida, S., Itoh, R. D., et al. (2024). Principles of amyloplast replication in the ovule integuments of Arabidopsis thaliana. Plant Physiol. 196(1): 137–152. https://doi.org/10.1093/plphys/kiae314
分类
植物科学 > 植物细胞生物学 > 细胞成像
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link