- EN - English
- CN - 中文
The Centriole Stability Assay: A Method to Investigate Mechanisms Involved in the Maintenance of the Centrosome Structure in Drosophila Cultured Cells
中心粒稳定性检测:研究果蝇培养细胞中中心体结构维持机制的方法
发布: 2025年06月05日第15卷第11期 DOI: 10.21769/BioProtoc.5330 浏览次数: 2212
评审: Vikash VermaAnonymous reviewer(s)
Abstract
Centrosomes are vital eukaryotic organelles involved in regulating cell adhesion, polarity, mobility, and microtubule (MT) spindle assembly during mitosis. Composed of two centrioles surrounded by the pericentriolar material (PCM), centrosomes serve as the primary microtubule-organizing centers (MTOCs) in proliferating cells. The PCM is crucial for MT nucleation and centriole biogenesis. Centrosome numbers are tightly regulated, typically duplicating once per cell cycle, during the S phase. Deregulation of centrosome components can lead to severe diseases. While traditionally viewed as stable structures, centrosomes can be inactivated or disappear in differentiating cells, such as epithelial cells, muscle cells, neurons, and oocytes. Despite advances in understanding centrosome biogenesis and function, the mechanisms maintaining mature centrosomes or centrioles, as well as the pathways regulating their inactivation or elimination, remain less explored. Studying centrosome maintenance is challenging as it requires the uncoupling of centrosome biogenesis from maintenance. Tools for acute spatial-temporal manipulation are often unavailable, and manipulating multiple components in vivo is complex and time-consuming. This study presents a protocol that decouples centrosome biogenesis from maintenance, allowing the study of critical factors and pathways involved in the maintenance of the integrity of these important cellular structures.
Key features
• Drosophila cultured cells are resistant to centriole reduplication during S phase arrest, making them a suitable model for studying centrosome integrity without confounding effects from centriole biogenesis.
Keywords: Centrosomes (中心体)Graphical overview
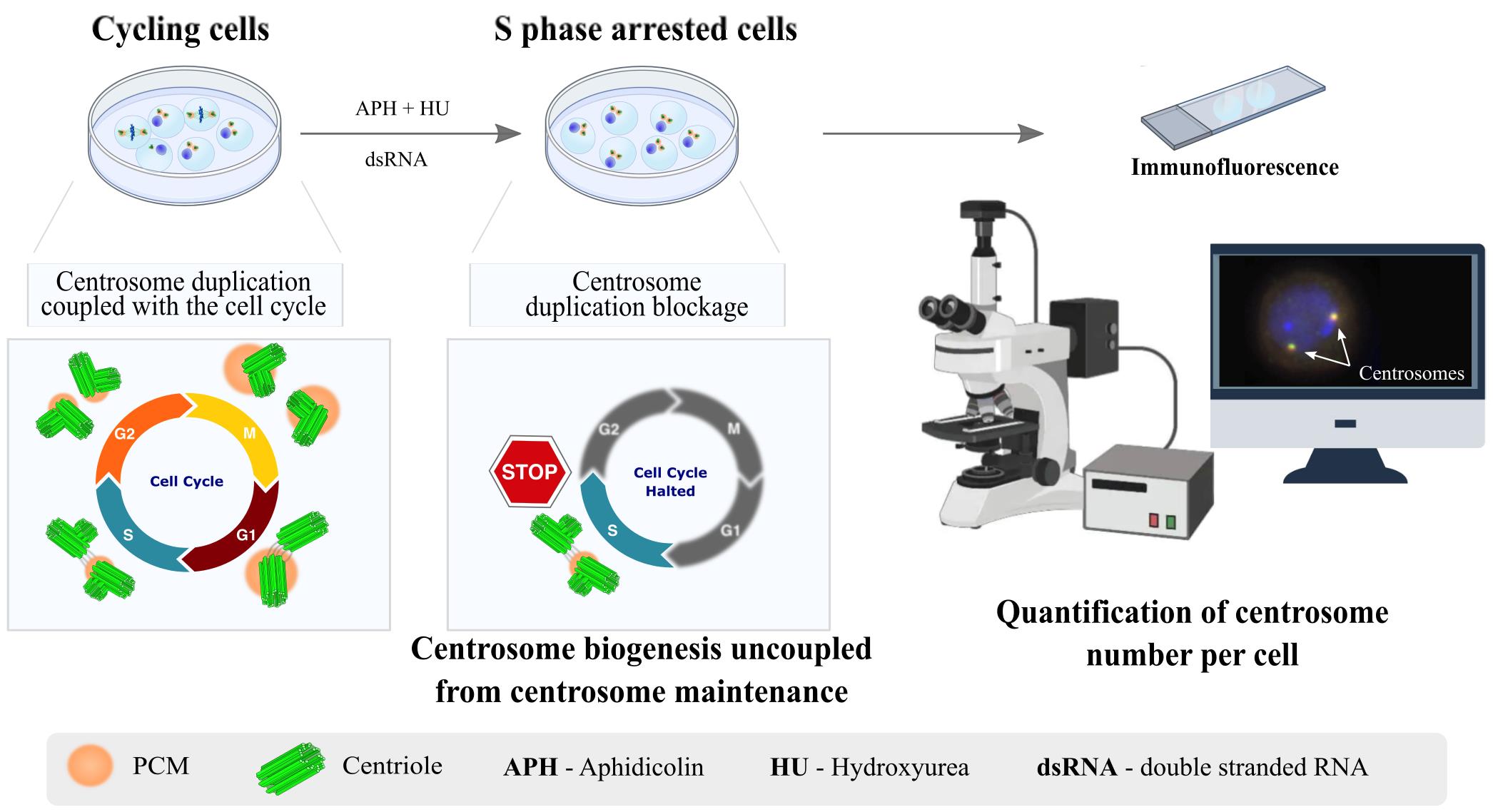
Background
An essential aspect of cell homeostasis is the maintenance of various cellular components, including proteins and lipids, and even the integrity of entire organelles. One such organelle is the centrosome, a conserved eukaryotic structure that plays a crucial role in several cellular processes. These include the regulation of cell adhesion, polarity, and mobility during interphase, as well as the assembly of the mitotic spindle, mainly composed of microtubules (MTs) [1,2]. Centrosomes serve as the primary microtubule-organizing centers (MTOCs) in proliferating eukaryotic cells. Each centrosome is composed of two centrioles—MT-based cylindrical structures surrounded by a multiprotein matrix known as the pericentriolar material (PCM) (Figure 1A). The PCM contains factors essential for MT nucleation and anchoring, also being critical for centriole biogenesis [3]. In cycling cells, the number of centrosomes is tightly controlled. Centrosomes are typically duplicated once per cell cycle, with duplication coupled to DNA replication, during the S phase [4]. In some cell types, during interphase, the centrosome migrates to the cell cortex, promoting the growth of an MT-based axoneme to form cilia or flagella [4]. Deregulation of centrosomal components leads to a wide range of diseases, many of which are incompatible with life.
Historically, centrosomes have been regarded as exceptionally stable structures [5,6]. However, in some differentiating cell types, these structures can be inactivated (i.e., lose their MTOC activity) or even disappear, as seen in epithelial cells, muscle cells, neurons, and oocytes [7–9]. Advances in various technological approaches have uncovered the major conserved components and pathways regulating centrosome duplication and maturation into fully functional MTOCs. However, much less is known about the components involved in the maintenance of mature centrosomes or centrioles, the pathways that regulate their inactivation or elimination, and how deregulation of these pathways may contribute to disease [10]. Hyperactive MTOC function at centrosomes has been linked to epithelial cancers and invasive tumor cell behavior [11–15]. Additionally, centrosome inactivation has been proposed as a strategy employed by cancer cells to silence supernumerary centrosomes, which would otherwise lead to cell death [16]. Therefore, elucidating the mechanisms that regulate centrosome and centriole stability is crucial not only for understanding the pathways of centrosome inactivation and/or elimination in post-mitotic and differentiating cells but also for uncovering how deregulation of these processes may contribute to cancer and other diseases.
Studying centrosome maintenance poses experimental challenges and limitations. Experimental setups must ensure that any manipulation (loss- or gain-of-function, such as in vivo RNAi or ectopic expression) is performed only after centrosomes have been fully assembled and that such manipulations do not affect centriole biogenesis. This requires tools for acute spatial-temporal manipulation, which are often unavailable or difficult to implement in many systems. Additionally, depending on the model system used, simultaneously manipulating (depleting or over-expressing) two or more components in vivo can be very challenging and time-consuming.
Taking into account these limitations, we provide a protocol that uncouples centriole biogenesis from maintenance in Drosophila melanogaster cultured cells (DMEL-2). By using this protocol, we have successfully identified components required for the maintenance of centrosome integrity [17,18]. That knowledge allowed us to generate tools to validate the function of such components in vivo [17,18]. In this assay, cells are arrested in S phase using aphidicolin (APH) and hydroxyurea (HU), which stall DNA replication. Unlike some commonly used human cultured cell lines, which allow for multiple rounds of centriole reduplication under S phase arrest conditions [19–21], Drosophila cultured cell lines do not exhibit this behavior. This distinction allows for the separation of centrosome duplication from centrosome maintenance. Moreover, Drosophila cultured cells are easy to culture and manipulate, and the addition of long double-stranded RNAs (dsRNA) enables the depletion of different proteins individually or simultaneously by RNAi. Therefore, following treatment with HU and APH, the number of centrioles is not affected by pathways involved in centriole biogenesis. This allows for the investigation of the effects of different experimental manipulations on centrosome stability.
Materials and reagents
Biological materials
1. Schneider’s Drosophila melanogaster cultured cell line 2 [D. Mel. (2), SL2] (DMEL-2, ATCC, catalog number: CRL-1963); cells isolated in 1969 from embryos
Reagents
1. Express 5 SFM medium (Gibco, catalog number: 10486025)
2. L-Glutamine (Thermo Scientific, catalog number: 25030024)
3. Penicillin-streptomycin-glutamine (PSG) (100×) (Gibco, catalog number: 10378016)
4. Aphidicolin (APH) (Sigma-Aldrich, catalog number: A0781)
5. Hydroxyurea (HU) (Sigma-Aldrich, catalog number: H8627)
6. Effectene Transfection Reagent kit (QIAGEN, catalog number: 301427)
7. Absolute methanol (Fisher Scientific, catalog number: 10675112)
8. 16% paraformaldehyde (formaldehyde) aqueous solution (Science Services GmbH, catalog number: E15710)
9. PIPES for buffer solutions (PanReac AppliChem, catalog number: A1079)
10. N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES) (1 M) (Gibco, catalog number: 15630080)
11. Ethylene glycol-bis (2-aminoethylether)-N,N,N',N'-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889)
12. Magnesium sulfate heptahydrate (MgSO4) (Sigma-Aldrich, catalog number: M1880)
13. Phosphate buffered saline (PBS) (Biowest, catalog number: L0615-500)
14. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912)
15. Triton X-100 (Sigma-Aldrich, catalog number: T8787)
16. Vectashield with DAPI mounting medium (Vector Laboratories, catalog number: H-1000-10)
Solutions
1. Express 5 SFM medium supplemented with L-glutamine (see Recipes)
2. Express 5 SFM medium supplemented with penicillin-streptomycin and L-glutamine (see Recipes)
3. Paraformaldehyde fixative solution (see Recipes)
4. PBSTB (see Recipes)
Recipes
1. Express 5 SFM medium supplemented with L-glutamine
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Express 5 SFM medium | n/a | 495 mL |
| 200 mM L-glutamine | 2 mM | 5 mL |
| Total | n/a | 500 mL |
Store at 4 °C.
2. Express 5 SFM medium supplemented with penicillin-streptomycin and L-glutamine
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Express 5 SFM medium | n/a | 495 mL |
| 100× PSG | 1× | 5 mL |
| Total | n/a | 500 mL |
Store at 4 °C.
3. Paraformaldehyde fixative solution
Prepare fresh before using. Cells in each well will require 400 μL of this fixative solution. Therefore, depending on the number of wells (conditions tested), different volumes of fixative solution might be required. Here, we present a recipe for 10 mL of fixative solution.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 0.2 M PIPES pH 6.8 | 60 mM | 3 mL |
| 0.2 M HEPES pH 7.0 | 30 mM | 1.5 mL |
| 0.5 M EGTA pH 6.8 | 10 mM | 200 μL |
| 1 M MgSO4 | 4 mM | 40 μL |
| 16% paraformaldehyde | 4% | 2.5 mL |
| ddH2O | n/a | 2.76 mL |
| Total | n/a | 10 mL |
4. PBSTB
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1× PBS | 1× | 99 mL |
| 10% Triton X-100 | 0.1% | 1 mL |
| BSA | 1% | 1 g |
| Total | n/a | 100 mL |
Prepare fresh and store at 4 °C.
Laboratory supplies
1. Tissue culture dish 150 mm (Sarstedt, catalog number: 3023421)
2. Cell culture flask, T-75 (Sarstedt, catalog number: 83.3911.002)
2. Cell culture flask, T-25 (Sarstedt, catalog number: 83.3910.002)
4. Tissue culture test plate, 6 wells (TPP, catalog number: 92006)
5. Tissue culture test plate, 24 wells (Corning, catalog number: 3526)
6. Round coverslips, 13 mm, No. 1.5 (VWR, catalog number: 631-0150)
7. Microscope slides (VWR, catalog number: 631-1553)
8. Hemocytometer (Neubauer)
9. Foil paper
Equipment
1. Cell culture flow hood
2. Cell culture incubator capable of maintaining a constant temperature of 25–27 °C
Note: The medium used to culture DMEL-2 cells does not require pH control; therefore, it is not necessary to use an incubator with a CO2 supply line.
3. Nikon Ti-E motorized microscope (computer-controlled microscope) equipped with a Zyla 4.2 Mpx sCMOS Andor camera (or equivalent with high quantum efficiency and low noise) and 100× 1.4 NA objective lens
Software and datasets
1. Fiji; Java-based program: https://imagej.net/software/fiji/ [22]
Procedure
文章信息
稿件历史记录
提交日期: Aug 26, 2024
接收日期: Apr 11, 2025
在线发布日期: May 16, 2025
出版日期: Jun 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Lince-Faria, M., Ferreira-Silva, A. and Pimenta-Marques, A. (2025). The Centriole Stability Assay: A Method to Investigate Mechanisms Involved in the Maintenance of the Centrosome Structure in Drosophila Cultured Cells. Bio-protocol 15(11): e5330. DOI: 10.21769/BioProtoc.5330.
分类
发育生物学 > 细胞生长和命运决定 > 分化
细胞生物学 > 细胞结构 > 微管
细胞生物学 > 细胞分离和培养
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













