- EN - English
- CN - 中文
Stable 13C-glutamine Tracing Resolved Metabolomics for Cancer Metabolism Study
稳定同位素¹³C-谷氨酰胺示踪代谢组学在癌症代谢研究中的应用
(*contributed equally to this work) 发布: 2025年05月20日第15卷第10期 DOI: 10.21769/BioProtoc.5322 浏览次数: 3214
评审: Elena A. OstrakhovitchTing MiaoAnonymous reviewer(s)
Abstract
Stable isotopes have frequently been used to study metabolic processes in live cells both in vitro and in vivo. Glutamine, the most abundant amino acid in human blood, plays multiple roles in cellular metabolism by contributing to the production of nucleotides, lipids, glutathione, and other amino acids. It also supports energy production via anaplerosis of tricarboxylic acid cycle intermediates. While 13C-glutamine has been extensively employed to study glutamine metabolism in various cell types, detailed analyses of specific lipids derived from 13C-glutamine via the reductive carboxylation pathway are limited. In this protocol, we present a detailed procedure to investigate glutamine metabolism in human glioblastoma (GBM) cells by conducting 13C-glutamine tracing coupled with untargeted metabolomics analysis using liquid chromatography–mass spectrometry (LC–MS/MS). The method includes step-by-step instructions for the extraction and detection of polar metabolites and long-chain fatty acids (LCFAs) derived from 13C-glutamine in GBM cells. Notably, this approach enables the distinction between isomers of two monounsaturated FAs with identical masses: palmitoleic acid (16:1n-7) (cis-9-hexadecenoic acid) and palmitelaidic acid (16:1n-7) (trans-9-hexadecenoic acid) derived from 13C-glutamine through the reductive carboxylation process. In addition, using this protocol, we also unveil previously unknown metabolic alterations in GBM cells following lysosome inhibition by the antipsychotic drug pimozide.
Key features
• Methods for analyzing the flux of the stable isotope 13C-glutamine in cancer cells and identifying its derived polar metabolites and long-chain fatty acids (LCFAs).
• Distinguishes isomers of long-chain fatty acids, such as palmitoleic acid (16:1n-7) (cis-9-Hexadecenoic acid) and palmitelaidic acid (16:1n-7) (trans-9-Hexadecenoic acid), which share the exact same mass.
• The method is utilized to investigate glutamine metabolism reprogramming in cancer cells following lysosome inhibition.
Keywords: 13C-glutamine (¹³C-谷氨酰胺)Graphical overview
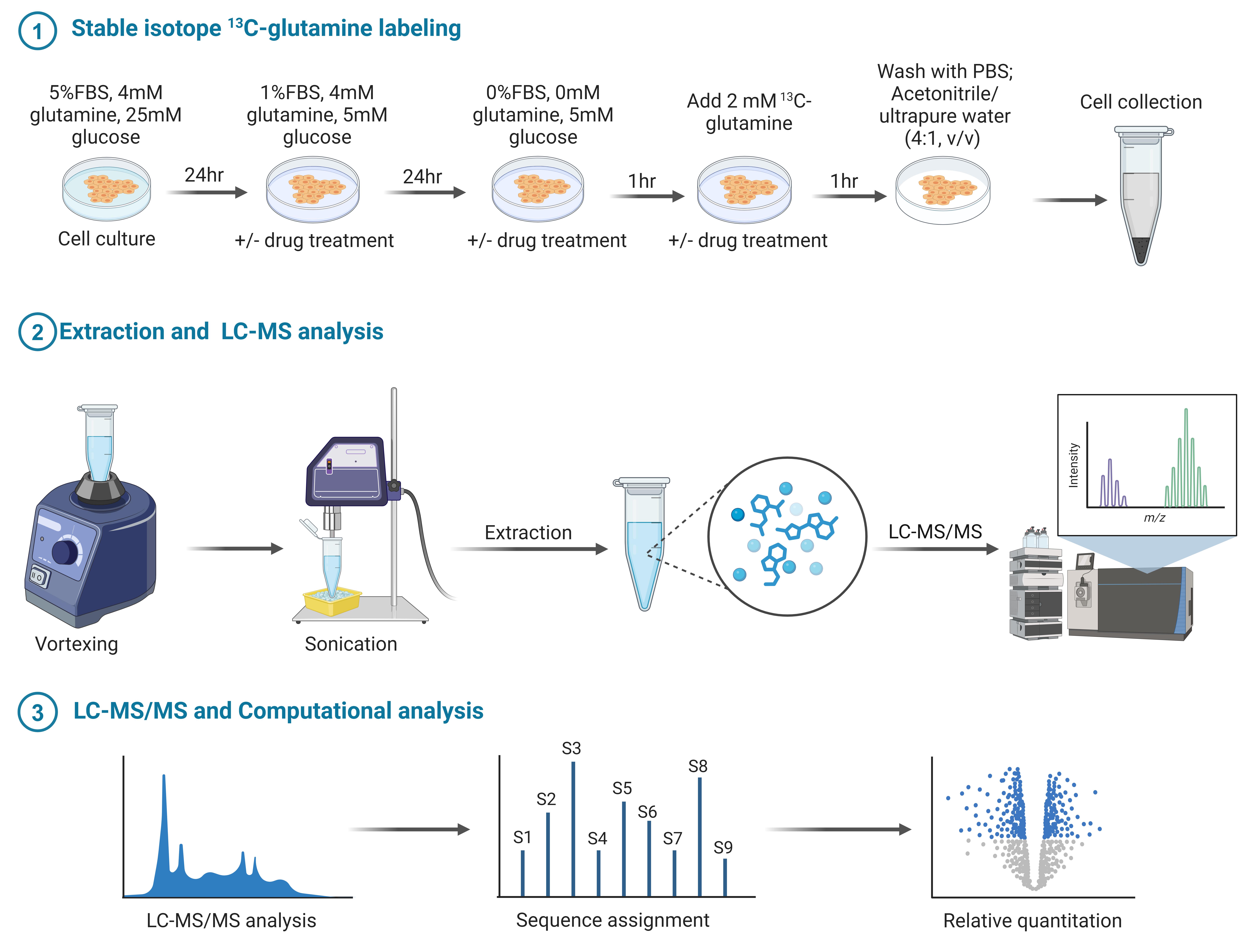
Metabolomic profiling of stable isotope 13C-glutamine labeling: Sample preparation and detection for polar metabolite and long-chain fatty acid (LCFA). Created with BioRender.com.
Background
Metabolism in live cells and tissues is a highly dynamic process, constantly changing in response to varying environmental conditions [1]. Conventional metabolomics studies, which measure metabolite levels at specific time points under defined conditions, fail to capture the dynamic nature of metabolic profiles, such as the active glucose and glutamine metabolism in different cell types [2–4]. In contrast, stable isotope 13C-labeling coupled with untargeted metabolomics analysis is a powerful tool for unveiling the comprehensive metabolic pathways originating from individual nutrients. For instance, 13C-glucose and 13C-glutamine are commonly used to study cellular metabolism both in vitro and in vivo. This approach has significantly advanced our understanding of metabolic processes in both physiological and pathological conditions [5–7]. However, most studies focus on water-soluble small molecular metabolites. The complexity of lipid structures has posed significant challenges for studying lipids derived from 13C-glucose or 13C-glutamine. There have been relatively few studies tracing long-chain fatty acids (LCFAs), such as palmitate (C:16), monounsaturated fatty acids including palmitoleic acid (16:1n-7) (cis-9-hexadecenoic acid), palmitelaidic acid (16:1n-7) (trans-9-hexadecenoic acid), and oleic acid (C:18:1), and polyunsaturated fatty acids like pinolenic acid (18:3) derived from 13C-glucose and 13C-glutamine, particularly in cancer cells. To bridge this research gap, we have developed a robust method to analyze the metabolic flux of 13C-glutamine into LCFAs in human glioblastoma (GBM) cells, the most lethal primary brain tumor cells [8].
Metabolic reprogramming is a hallmark of aggressive cancers, including GBM [9–13]. Recent studies have demonstrated that both glutamine metabolism and de novo lipid synthesis are highly upregulated in various cancers [9]. However, the underlying connection between these two crucial metabolic pathways remains poorly understood. Our recent studies, published in Nature Metabolism (2022) [9] and Cell Reports Medicine (2024) [8], revealed that glutamine, through its released ammonia, directly activates lipid synthesis to promote GBM growth. Furthermore, we successfully developed a protocol to examine the flux of glutamine conversion into LCFAs [8]. Using 13C-glutamine tracing studies, we uncovered metabolic responses of GBM cells to lysosome inhibition [8]. Specifically, we identified that the antipsychotic drug pimozide enters lysosomes and inhibits their function, revealing new therapeutic possibilities [8]. This method is not limited to GBM cells; it can be applied to various other cell types to investigate glutamine metabolism, particularly for determining the extent of LCFA synthesis derived from glutamine. These findings provide a valuable platform for understanding and targeting metabolic pathways in cancer.
Materials and reagents
Biological materials
1. Human GBM cell line U251 (Sigma-Aldrich, catalog number: 09063001) [8,11,12]
Reagents
1. Dulbecco’s modified Eagle’s medium (DMEM), no glutamine (Corning, catalog number: 15-0312CV) (for regular cell culture)
2. Dulbecco’s modified Eagle’s medium (DMEM), no glucose, no glutamine, no phenol red (Gibco, catalog number: A14430-01) (used during drug treatment)
3. L-glutamine (12C-Glutamine) (Gibco, catalog number: A2916801); prepare at 200 mM, aliquot, and store at -80°C until use
4. 13C-glutamine (Sigma, catalog number: 184161-19-1); dissolve in PBS to a final concentration of 200 mM, aliquot, and store at -80°C until use
5. Glucose (Sigma, catalog number: G8644)
6. Sodium pyruvate (Gibco, catalog number: 11360070)
7. Fetal bovine serum (FBS) (Cytiva, catalog number: SH30071.03)
8. Cell culture phosphate buffered saline (PBS) (Corning, catalog number: 21-040); store in a 4 °C refrigerator overnight before use
9. Dimethyl sulfoxide (DMSO) (Sigma, catalog number: D2650)
10. Trypan blue (Sigma, catalog number: T8154) (for cell counting)
11. HyCloneTM HyPure water, molecular biology grade (Cytiva, catalog number: SH30538.02); store in a 4 °C refrigerator overnight before use
12. Acetonitrile (CH3CN or ACN) (Fisher Chemical, catalog number: A955); place a bottle of 100% CH3CN (HPLC grade) in a -20 °C freezer overnight prior to use
13. Chloroform (CHCl3) (Sigma, catalog number: C2432)
14. Methanol (Fisher Chemical, catalog number: A412)
15. Ethanol (Sigma, catalog number: E7023)
16. Acetic acid (Sigma, catalog number: 695092)
17. Pimozide (Sigma, catalog number: P1793); dissolve in DMSO to a final concentration of 10 mM, aliquot, and store at -80°C until use
18. Deionized water (diH2O) (VION Biosciences, catalog number: DH2O-10).
Laboratory supplies
1. Cell culture dish, 100 mm (Alkali Scientific, catalog number: TDN0101)
2. Cell culture dish, 150 mm (Alkali Scientific, catalog number: TDN0150)
3. 1,250 μL pipette tips (Alkali Scientific, catalog number: RT1250)
4. 200 μL pipette tips (Alkali Scientific, catalog number: RT1200)
5. 10 μL pipette tips (Alkali Scientific, catalog number: RT1010)
6. 1.5 mL Eppendorf tubes (Alkali Scientific, catalog number: C3016-S)
7. Cell scraper (Fisher brand, catalog number: 08100242)
8. 15 mL screw-cap conical tube (Alkali Scientific, catalog number: CN5600)
9. Liquid nitrogen
10. Tube rack
11. Ice
12. Ice box
13. SureSTART LC autosampler vial (Thermo Fisher, catalog number: 03-452-235)
14. OASIS HLB cartridge (Waters Corp., catalog number: WAT094225)
15. Leap PAL brand cap (Fisher Scientific, catalog number: 50-125-0992)
Equipment
1. CellDrop FL cell counter (DeNovix, USA)
2. Cell culture incubator (VWR International, USA)
3. Inverted microscope (Fisher Scientific, USA)
4. P1000 pipette (Gilson Incorporated, USA)
5. P200 pipette (Gilson Incorporated, USA)
6. P10 pipette (Gilson Incorporated, USA)
7. P2 pipette (Gilson Incorporated, USA)
8. Vortex (Fisher Scientific, USA)
9. Sonic Dismembrator (Fisher Scientific, USA)
10. Thermo Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA)
11. Thermo DIONEX UltiMate 3000 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA)
12. Centrifuge 5804 R (Eppendorf, Enfield, CT, USA)
13. Microfuge 22R centrifuge (Beckman Coulter, Brea, CA, USA)
14. FreeZone 2.5 plus (LABCONCO, Kansas, MO, USA)
Software and datasets
1. XCMS software (https://xcmsonline.scripps.edu/landing_page.php?pgcontent=mainPage) for LC–MS data deconvolution
2. MetSign software for metabolite cross-sample peak list alignment and normalization
3. Compound Discoverer software (v 2.0, Thermo Fisher Scientific, Germany) for metabolite identification from both the in-house database and public database
4. BioRender.com was used for the graphical overview
5. National Metabolomics Data Repository (NMDR), the Metabolomics Workbench (https://www.metabolomicsworkbench.org/) (access date, 12-12-2024)
6. All data and code have been deposited to NMDR: https://www.metabolomicsworkbench.org/ (access date, 12-12-2024)
Procedure
文章信息
稿件历史记录
提交日期: Jan 3, 2025
接收日期: Apr 26, 2025
在线发布日期: May 8, 2025
出版日期: May 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Zhong, Y., He, L., Yin, X., Mazik, L., Zhang, X. and Guo, D. (2025). Stable 13C-glutamine Tracing Resolved Metabolomics for Cancer Metabolism Study. Bio-protocol 15(10): e5322. DOI: 10.21769/BioProtoc.5322.
分类
癌症生物学 > 癌症生物化学 > 癌代谢
系统生物学 > 代谢组学 > 组织
生物化学 > 脂质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













