- EN - English
- CN - 中文
From Bedside to Desktop: A Data Protocol for Normative Intracranial EEG and Abnormality Mapping
从床旁到桌面:标准化颅内脑电与异常图谱的数据处理方案
发布: 2025年05月20日第15卷第10期 DOI: 10.21769/BioProtoc.5321 浏览次数: 1762
评审: Xiaochen SunOneil Girish BhalalaSébastien Gillotin
Abstract
Normative mapping is a framework used to map population-level features of health-related variables. It is widely used in neuroscience research, but the literature lacks established protocols in modalities that do not support healthy control measurements, such as intracranial electroencephalograms (icEEG). An icEEG normative map would allow researchers to learn about population-level brain activity and enable the comparison of individual data against these norms to identify abnormalities. Currently, no standardised guide exists for transforming clinical data into a normative, regional icEEG map. Papers often cite different software and numerous articles to summarise the lengthy method, making it laborious for other researchers to understand or apply the process. Our protocol seeks to fill this gap by providing a dataflow guide and key decision points that summarise existing methods. This protocol was heavily used in published works from our own lab (twelve peer-reviewed journal publications). Briefly, we take as input the icEEG recordings and neuroimaging data from people with epilepsy who are undergoing evaluation for resective surgery. As final outputs, we obtain a normative icEEG map, comprising signal properties localised to brain regions. Optionally, we can also process new subjects through the same pipeline and obtain their z-scores (or centiles) in each brain region for abnormality detection and localisation. To date, a single, cohesive dataflow pipeline for generating normative icEEG maps, along with abnormality mapping, has not been created. We envisage that this dataflow guide will not only increase understanding and application of normative mapping methods but will also improve the consistency and quality of studies in the field.
Key features
• Resultant normative maps can be used to test a broad range of hypotheses in the neuroscience field.
• Provides a more detailed walkthrough of the methods in the normative mapping study conducted by Taylor et al. [1] and other related publications [2–12].
• Offers flexibility: readers can tailor the final output by considering key decision points included throughout the protocol.
• Involves sub-pipelines, which may be useful to researchers in isolation (i.e., icEEG electrode localisation and/or interictal segment selection).
Keywords: Data pipeline (数据流程)Graphical overview
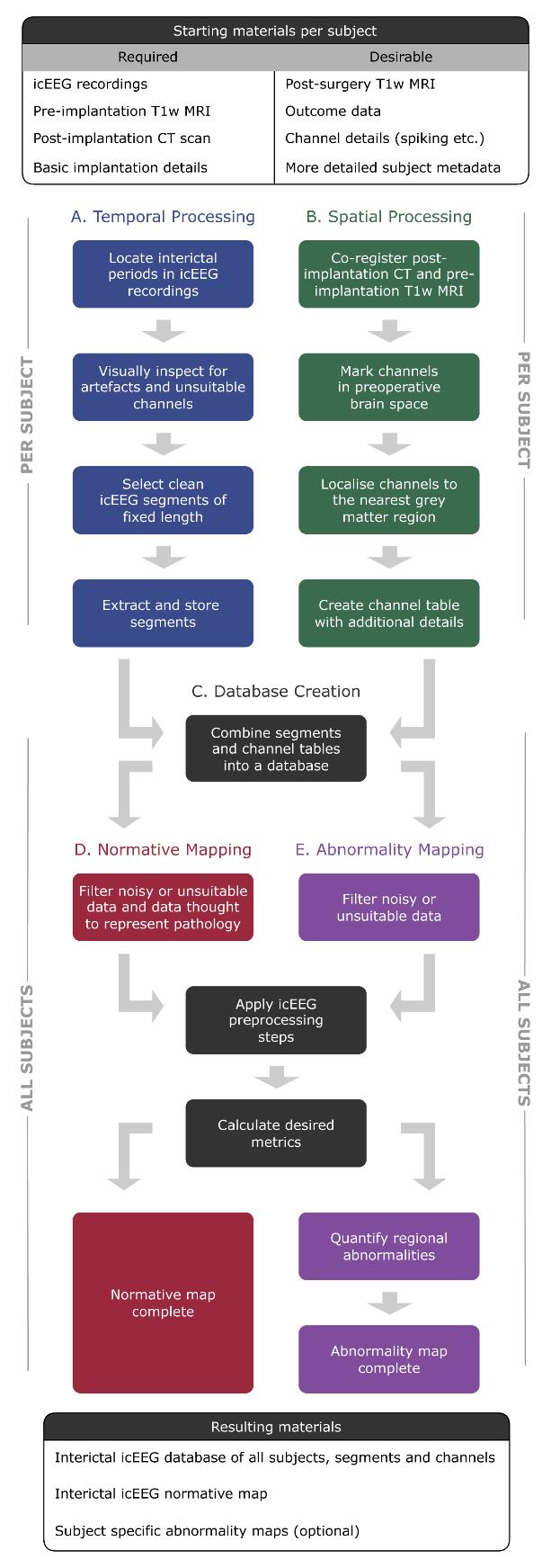
Flowchart of pipelines involved in this protocol. Temporal processing (pipeline A) and spatial processing (pipeline B) can be run sequentially or in parallel.
Background
Normative mapping is a framework in which features of health-related variables are mapped at the population level. A common example is growth charts, used to evaluate whether a child’s growth falls within the normal range for their age. In neuroscience research, this framework is widely used to map normative ranges of brain activity, for example.
However, normative mapping is particularly challenging in modalities that do not support measurements in healthy control subjects, such as the highly invasive intracranial electroencephalogram (icEEG). An icEEG normative map of brain activity is valuable because it allows us to better understand populations of interest. Further, individuals can be compared to the normative map, helping to identify, characterise, and localise potentially pathological abnormalities. Such work is available in the literature [1–7,13–20], revealing, for example, that icEEG normative mapping can localise epileptogenic tissue [1], and that the abnormality maps that follow are temporally stable [5].
Despite these promising findings, there is currently no detailed guide available for transforming clinical data, such as neuroimaging and icEEG recordings, into a normative map of brain activity localised to standardised brain regions. This literature gap hinders the consistent application of the methodology across studies. To remedy this, our protocol consolidates existing methods and provides key decision points for constructing normative maps comprising regional signal properties, along with (optionally) regional abnormality maps for new subjects.
One of the protocol’s strengths lies in its modular design. Distinct sub-pipelines, such as the spatial processing pipeline, offer a standalone utility. For instance, our own lab has employed this pipeline to demonstrate that the incomplete resection of the icEEG seizure onset zone (SOZ) is not associated with post-surgical outcomes [10], amongst other results [8,9,11,12]. Furthermore, this protocol is written as a dataflow guide, allowing for flexibility. Researchers can choose their preferred programming language and data management software for implementation, decide whether to compute abnormalities, and apply the temporal processing pipeline to other modalities, such as scalp EEG (for instance). Our protocol allows for adaptation to best suit the reader.
While the protocol provides a standardised framework, certain aspects, such as manual resection mask delineation, could benefit from modernisation. Automated methods exist within our own lab [21] and externally [22–25], although their reliability varies [26]. The flexible, guide-like nature of our protocol is both a strength and a weakness. Importantly, we offer pseudo code and emphasise dataflow over low-level implementation, allowing researchers to tailor the process to their needs while fostering a deeper understanding of the method and related literature.
This protocol provides a cohesive data flow on icEEG normative and abnormality mapping, which is currently missing from the literature. We aim to equip neuroscience researchers with the tools needed to develop normative maps, allowing them to explore new hypotheses consistently and effectively.
Equipment
1. PC or laptop capable of running the required software (below) with sufficient storage space. As a rule of thumb, we recommend at least 1 GB per subject. As an example, a standard Macbook Pro would be capable of running the full protocol. Increasing computational resources would allow a larger number of subjects to be processed more efficiently.
Software and datasets
Required
1. icEEG recordings for each subject
2. Post-implantation computed tomography (CT) scan for each subject
3. Pre-implantation volumetric T1-weighted magnetic resonance imaging (T1w MRI) for each subject
4. Programming environment (this is the reader’s choice, but common examples include MATLAB, Python, and R; see the General Notes section)
5. Software to view icEEG data (e.g., EDFBrowser or the reader’s chosen programming environment)
6. FreeSurfer [27] (we used version 7.3)
7. Electrode localisation tools such as img_pip [28]
8. Database software (e.g., MongoDB)
Optional/recommended
9. Post-surgery T1w MRI for subjects who proceeded to resective epilepsy surgery
10. ANTS toolbox [29]
11. 3D image viewer such as FSLeyes or ITK-Snap
12. RAMPS pipeline [21]
Notes:
1. If the reader is looking for suitable data, some of our published works [1–7] use the publicly available RAM dataset, found at https://memory.psych.upenn.edu/RAM.
2. Some of the above software may have certain CPU or license requirements. Please refer to each tool's documentation and/or official website page for the most up-to-date information.
3. We have included a glossary in the General notes section to avoid any ambiguity around our chosen vocabulary, e.g., electrode contact and electrode channel are often used interchangeably in the literature.
Prerequisites
The experience of the reader should be high for programming, data organisation, neuroimaging, and EEG signal processing.
Procedure
文章信息
稿件历史记录
提交日期: Feb 6, 2025
接收日期: Apr 15, 2025
在线发布日期: May 13, 2025
出版日期: May 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Woodhouse, H., Gascoigne, S. J., Hall, G., Simpson, C., Evans, N., Schroeder, G. M., Taylor, P. N. and Wang, Y. (2025). From Bedside to Desktop: A Data Protocol for Normative Intracranial EEG and Abnormality Mapping. Bio-protocol 15(10): e5321. DOI: 10.21769/BioProtoc.5321.
分类
生物信息学与计算生物学
神经科学 > 神经系统疾病 > 癫痫
神经科学 > 基础技术 > 脑电图
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













