- EN - English
- CN - 中文
Mycobacterium smegmatis Ribosome Purification, Co-sedimentation, and Subunit Association Assay
解脲分枝杆菌核糖体的纯化、共沉淀及亚基重组分析方法
发布: 2025年05月20日第15卷第10期 DOI: 10.21769/BioProtoc.5318 浏览次数: 1581
评审: Ritu GuptaAnonymous reviewer(s)
Abstract
The ribosome, a complex macromolecular machine, plays a vital role in cellular translation. To investigate its structure and conduct in vitro experiments, isolating the ribosomes from cells is the first step. While isolating ribosomes from bacterial cells is routine, obtaining them from mycobacteria proves challenging due to the protective mycolic acid layer, which hinders cell lysis. In this study, we present a straightforward and efficient protocol for isolating ribosomes from Mycobacterium smegmatis. Additionally, we introduce a co-sedimentation assay using density gradient ultracentrifugation, providing a simple yet powerful method for studying ribosome–protein interactions. The re-association assay also offers a practical approach for obtaining tRNA-free 70S ribosomes and evaluating the anti-association properties of potential ligands. While these assays are commonly used, our protocol stands out for its simplicity, requiring limited specialized instruments. These methods can also be scaled up or down per requirement. By employing sonication for cell rupture and utilizing basic lab equipment for ultracentrifugation-based assays, our method greatly simplifies ribosome isolation and related research.
Key features
• Cellular ribosome isolation from Mycobacterium smegmatis using an optimized sonication cycle.
• Detection of ribosome–protein interactions through density gradient ultracentrifugation.
• Simple steps to obtain tRNA-free 70S ribosomes that are crucial for several biochemical experiments.
• Screening of proteins or ligands for their ribosomal anti-association activity.
Keywords: 70S ribosome (70S核糖体)Graphical overview
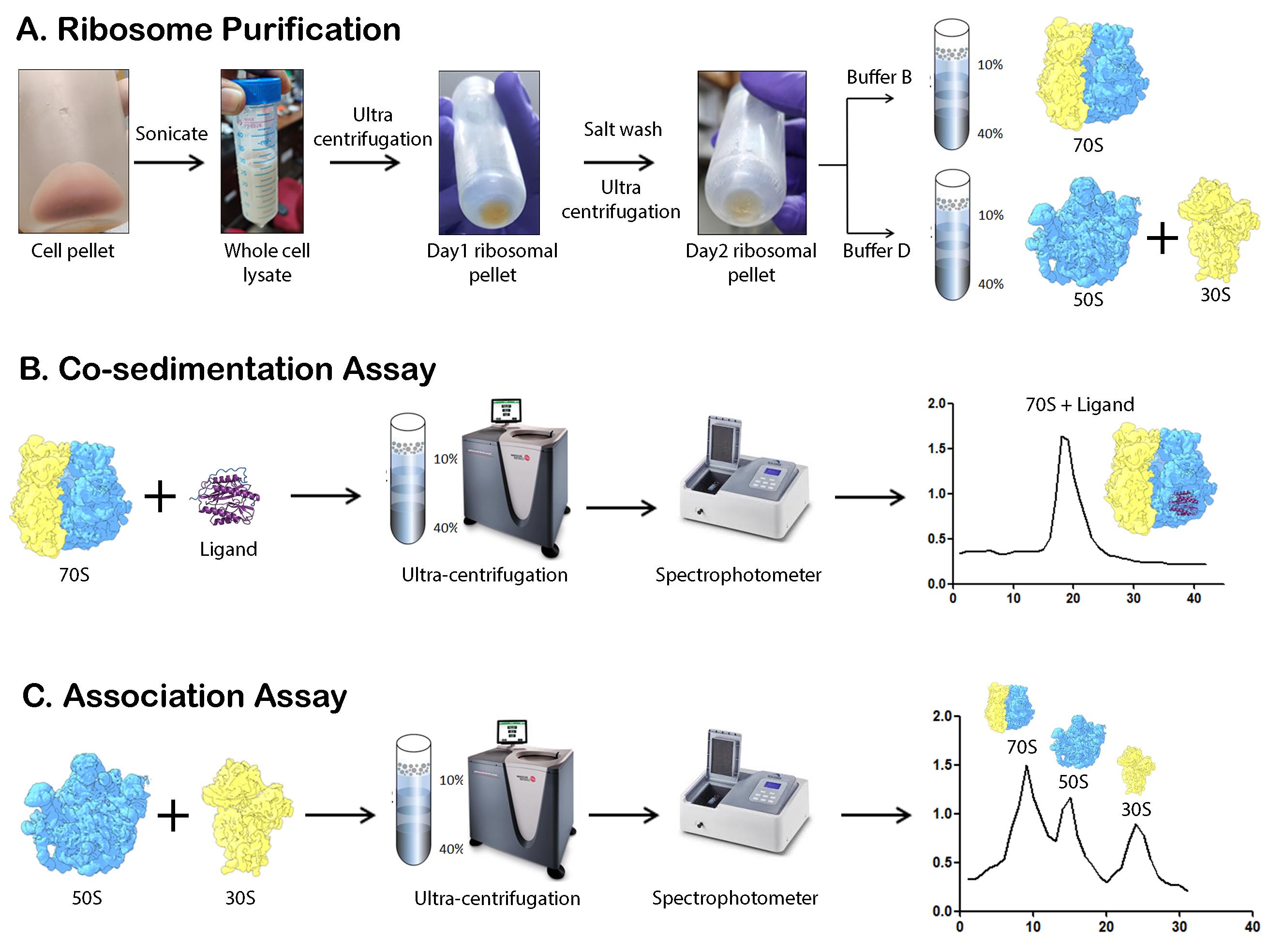
Schematic overview of the major steps involved in M. smegmatis ribosome purification, co-sedimentation, and association studies. (A) The schematic representation highlights the major steps involved in 70S, 50S, and 30S subunit isolation. Cells are lysed by sonication followed by two rounds of ultracentrifugation to obtain the crude ribosome. Depending on the requirement, the pellet is either dissolved in Buffer B or Buffer D, followed by sucrose density gradient ultracentrifugation. (B) Co-sedimentation assay involves the incubation of the ligand with the ribosome and analysis by ultracentrifugation. (C) Association assay involves the incubation of the 50S and 30S ribosomal subunits, followed by ultracentrifugation to analyze the amount of 70S ribosome formed.
Background
In all living cells, protein synthesis is carried out by the ribosome, a large complex composed of RNA and proteins [1–3]. Bacterial ribosomes are composed of a small 30S subunit (SSU) and a large 50S subunit (LSU). Structural studies of ribosomes from both Gram-negative and Gram-positive species have shown that the overall structure of the bacterial ribosome is largely conserved [4]. In E. coli, the 30S subunit consists of 21 ribosomal proteins (r-proteins) and a 16S ribosomal RNA (rRNA), while the 50S subunit contains 36 r-proteins along with 23S and 5S rRNAs. While the core of the ribosome is almost fully conserved, there are subtle variations in ribosome structure. These include species-specific insertions and deletions in various domains of rRNA and r-proteins, as well as novel r-proteins in certain bacterial species [5].
Ribosome isolation is a fundamental step in studying translation, ribosome structure, and ribosome-associated interactions [6]. Mycobacterium is an important bacterial genus that encompasses several prominent pathogens, and its translation machinery is of immense interest due to its implications in antibiotic approaches. While ribosome purification from bacterial cells is well-established, mycobacteria present unique challenges due to their thick, lipid-rich mycolic acid layer, which complicates cell lysis and downstream isolation procedures [7, 8]. Traditional methods often rely on harsh chemical and physical treatments (freeze-thaw), which can be time-consuming and costly, may compromise ribosome integrity, and may result in low yields. Previous studies have utilized various approaches, including mechanical disruption, detergent-based lysis, and enzymatic treatments [9,10], to extract ribosomes from mycobacteria. However, these methods often require specialized reagents, prolonged processing times, or multiple purification steps, making them less accessible for routine laboratory use.
Our protocol offers a simplified yet efficient approach, using sonication for cell disruption and basic ultracentrifugation techniques without the need for alcohol-based precipitation or specialized equipment. This method may be adapted to other mycobacterium species, provided proper safety precautions are taken. The method enables the isolation of high-quality ribosomes suitable for structural and functional studies, including ribosome–protein interaction assays via co-sedimentation as well as functional assays using re-association assays. This protocol has broader applications, such as screening ribosome-targeting antibiotics, studying translation regulation, and investigating ribosome-associated stress responses in mycobacteria. These methods may be scaled up or scaled down as per requirement. Its simplicity and accessibility make it a valuable tool for researchers exploring bacterial ribosome dynamics in both fundamental and applied sciences.
Materials and reagents
Biological materials
1. Mycobacterium smegmatis MC2 155 (gift from Prof. Sujoy Dasgupta, Bose Institute, Kolkata)
Reagents
1. Tris base (Fisher Scientific, catalog number: BP152-1)
2. Magnesium acetate (C4H6MgO4·4H2O) (Sigma-Aldrich, catalog number: M5661-250G)
3. Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: A9434-1KG)
4. 2-Mercaptoethanol (C2H6OS) (Sigma-Aldrich, catalog number: M-7154)
5. Sucrose (C12H22O11) (Calbiochem, catalog number: 8510)
6. HEPES (Fisher Scientific, catalog number: BP310-1)
7. Middlebrook 7H9 broth base (Himedia, catalog number: M198-500G)
8. Bovine serum albumin (SRL, catalog number: 9048-46-8)
9. Polysorbate 80 (SRL, catalog number: 9005-65-6)
10. Glycerol (Sigma-Aldrich, catalog number: G5516-1L)
11. HCl (Rankem, catalog number: H0100)
12. Puromycin (Sigma-Aldrich, catalog number: P8833)
13. Lysozyme (Roche, catalog number: 10837059001)
14. Bis-acrylamide (30%) (Himedia, catalog number: ML037)
15. TEMED (SRL, catalog number: 52145)
16. Ammonium persulphate (SRL, catalog number: 0144142)
17. Sodium lauryl sulphate (SRL, catalog number: 32096)
18. Agarose (SRL, catalog number: 23287)
19. 50× TAE (G2P, catalog number: BUF-07-500ml)
20. Sodium hypochlorite (Sigma-Aldrich, catalog number: S-1898)
Solutions
1. Buffer A (see Recipes)
2. Buffer B (see Recipes)
3. Buffer C (see Recipes)
4. Buffer D (see Recipes)
5. HMA-10 (see Recipes)
6. Mycobacterium smegmatis MC2 155 culture media (see Recipes)
7. Resolving and stacking gel for SDS-PAGE (see Recipes)
8. Agarose gel (see Recipes)
9. Sucrose solutions (see Recipes)
10. Puromycin stock (see Recipes)
Recipes
Note: The different types of buffers used in the experiments are mentioned in the tables below. All stocks are prepared using distilled water, and the final volume is adjusted using the same water. Buffers are then filtered using 0.22 μm membrane syringe filters. The filtered buffers can be stored at 4 °C for a few weeks, although it is ideal to use them within a week. Culture media is prepared using distilled water and autoclaved.
1. Buffer A
| Reagent | Stock concentration | Final concentration | Volume for 500 mL |
|---|---|---|---|
| Tris-HCl (pH 7.6) | 1 M | 20 mM | 10 mL |
| Ammonium chloride | 2 M | 100 mM | 25 mL |
| Magnesium acetate | 1 M | 10 mM | 5 mL |
| 2-Mercaptoethanol | 14.3 M | 5 mM | 175 μL |
2. Buffer B
| Reagent | Stock concentration | Final concentration | Volume for 500 mL |
|---|---|---|---|
| Tris-HCl (pH 7.6) | 1 M | 20 mM | 10 mL |
| Ammonium chloride | 2 M | 30 mM | 7.5 mL |
| Magnesium acetate | 1 M | 10 mM | 5 mL |
| 2-Mercaptoethanol | 14.3 M | 5 mM | 175 μL |
3. Buffer C
| Reagent | Stock concentration | Final concentration | Volume for 500 mL |
|---|---|---|---|
| Tris-HCl (pH 7.6) | 1 M | 20 mM | 10 mL |
| Ammonium chloride | 2 M | 1 M | 250 mL |
| Magnesium acetate | 1 M | 10 mM | 5 mL |
| 2-Mercaptoethanol | 14.3 M | 5 mM | 175 μL |
4. Buffer D
| Reagent | Stock concentration | Final concentration | Volume for 500 mL |
|---|---|---|---|
| Tris-HCl (pH 7.6) | 1 M | 20 mM | 10 mL |
| Ammonium chloride | 2 M | 30 mM | 7.5 mL |
| Magnesium acetate | 1 M | 1 mM | 0.5 mL |
| 2-Mercaptoethanol | 14.3 M | 5 mM | 175 μL |
5. HMA-10
| Reagent | Stock concentration | Final concentration | Volume for 500 mL |
|---|---|---|---|
| HEPES (pH 7.6) | 1 M | 20 mM | 10 mL |
| Magnesium acetate | 1 M | 10 mM | 5 mL |
| Ammonium chloride | 2 M | 60 mM | 15 mL |
| 2-Mercaptoethanol | 14.3 M | 5 mM | 175 μL |
6. Mycobacterium smegmatis MC2 155 culture media
| Reagent | For 1,000 mL |
|---|---|
| Middlebrook 7H9 broth base | 5.3 g |
| Bovine serum albumin | 1 g |
| Polysorbate 80 | 200 μL |
| Glycerol | 2 mL |
7. Resolving and stacking gel for SDS-PAGE
| Reagents for 15% resolving gel | Amount (for 10 mL) |
|---|---|
| Distilled water | 2.2 mL |
| Bis-acrylamide (30%) | 5 mL |
| 1.5 M Tris (PH 8.8) | 2.6 mL |
| 10% SDS | 100 μL |
| 10% APS | 100 μL |
| TEMED | 10 μL |
| Reagents for 5% stacking gel | Amount (for 5 mL) |
| Distilled water | 2.975 mL |
| Bis-acrylamide (30%) | 0.67 mL |
| 1 M Tris (pH 6.8) | 1.25 mL |
| 10% SDS | 50 μL |
| 10% APS | 50 μL |
| TEMED | 5 μL |
8. Agarose gel
| Reagent | Amount (for 100 mL) |
|---|---|
| Agarose | 1.2 g |
| 50× TAE | 2 mL |
| Sodium hypochlorite | 1 mL |
| Distilled water | 97 mL |
9. Sucrose solutions
| Amount of sucrose | Volume of buffer | Final % of sucrose |
|---|---|---|
| 4.8 g | 12 mL | 40% |
| 2.4 g | 8 mL | 30% |
| 1.4 g | 7 mL | 20% |
| 0.6 g | 6 mL | 10% |
10. Puromycin stock
| Puromycin | Buffer D | Final concentration |
|---|---|---|
| 0.0013 g | 100 µL | 13 mg/mL |
Laboratory supplies
1. Pipette (1 mL, 100 μL, 10 μL, and 2.5 μL) (Eppendorf, catalog number: 05-403-151)
2. Microcentrifuge tubes (1.5 mL) (Abdos, catalog number: P10202)
3. 0.22 μm syringe filters (Merck, catalog number: SLGB033RS)
4. 2 L Erlenmeyer narrow mouth conical flask (Borosil, catalog number: 4980030)
5. 50 mL centrifuge tubes (Abdos, catalog number: P10431)
6. 500 mL culture harvest bottles (Nalgene® bottles) (Sigma-Aldrich, catalog number: B9282)
7. Glass homogenizer (Potter-Elvehjem PTFE pestle and glass tube) (Sigma-Aldrich, catalog number: P7984)
8. Oakridge tubes (Nalgene® centrifuge tubes) (Sigma-Aldrich, catalog number: T1418)
9. Centrifuge tubes for ultracentrifugation (Beckman, catalog number: 344058)
10. Amicon® Ultra-4 centrifugal filters, 100 kDa and 10 kDa (Merck, catalog numbers: UFC810024 and UFC801024)
11. Measuring cylinder (Tarsons, catalog number: 343070)
12. Non-absorbent cotton (Himedia, catalog number: LA1017)
13. Quartz cuvette 1 mL (BR Biochem, catalog number: BQ-244)
14. 1 mL pipette tips (Abdos, catalog number: P10114)
15. 200 μL pipette tips (Abdos, catalog number: P10149)
16. 10 μL pipette tips (Abdos, catalog number: P10117)
17. Parafilm (Amcor, catalog number: PM-996)
Equipment
1. Crude weighing balance (Hexane, India, model: DJ602A)
2. Fine weighing balance (Mettler Toledo, model: ME204)
3. Magnetic stirrer (Spinot)
4. Autoclave (Indfos, Autoclave)
5. Class I laminar airflow system (custom assembled)
6. Incubator shaker (Remi, model: CIS-24 Plus)
7. Spectrophotometer (Shimadzu Corp, model: A114546)
8. 4 °C centrifuge (Sigma, model: 330k)
9. Sonicator (Qsonica Sonicators, model: Q125)
10. Ultracentrifuge with AH-629 swing-out rotor (Thermo Scientific, model: Sorval Max Ultra Series)
11. Vortex (Spinix, model: 3737)
12. Peristaltic pump (GE Healthcare Biosciences AV, model: PumpP-1)
13. 4 °C refrigerator (Vestfrost Solutions)
14. -20 °C freezer (Vestfrost)
Software and datasets
1. Prism (v5.00) software (GraphPad, San Diego, CA)
Note: License is required and can be purchased from the website. Graphs can also be prepared using Microsoft Excel.
2. UVProbe 2.42
Note: The license comes with the spectrophotometer.
Procedure
文章信息
稿件历史记录
提交日期: Mar 3, 2025
接收日期: Apr 18, 2025
在线发布日期: May 8, 2025
出版日期: May 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Banerjee, A., Dey, S., Srinivasan, K., Dhur, A., Baid, P. and Sengupta, J. (2025). Mycobacterium smegmatis Ribosome Purification, Co-sedimentation, and Subunit Association Assay. Bio-protocol 15(10): e5318. DOI: 10.21769/BioProtoc.5318.
分类
微生物学 > 微生物细胞生物学 > 细胞器分离
生物化学 > RNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













