- EN - English
- CN - 中文
I-PREFR: Inverse PCR-Based Restriction Enzyme FRee Unidirectional Strategy for Rapid Markerless Chromosomal Gene Deletion and Reconstitution in Bacteria Using Suicide Vectors
I-PREFR:基于反向PCR的无酶单向策略,利用自杀载体在细菌中快速实现无标记染色体基因缺失与重建
发布: 2025年05月20日第15卷第10期 DOI: 10.21769/BioProtoc.5314 浏览次数: 2367
评审: Emilia KrypotouJoyce ChiuDamián Lobato-Márquez
Abstract
The standard protocols for allelic exchange using homologous recombination deploy suicide vectors with negative selection markers. However, the use of multiple restriction enzymes to generate sticky ends in the vector and the insert for cloning is time-consuming, resource-intensive, and challenging. The advent of next-generation proofreading enzymes is enabling researchers to routinely carry out long-range PCR. Hence, amplifying 5–6 kb of complete low-complex DNA cloning vectors and 2–3 kb of complex genomic regions is much easier. Here, we report a simple, accurate, rapid, and unidirectional approach for chromosomal in-frame gene deletion and complementation by reconstitution of the full-length gene without using any restriction enzymes. The method requires long-range PCR using Phusion polymerase to linearize the vector and amplify the target gene to create a recombinant vector (pRM1) and further inverse PCR amplification of pRM1 to create a recombinant vector (pRM4) with a deleted version of the gene. The cloning steps involve the use of kinase and ligase for phosphorylation and ligation steps, respectively. The recombinant plasmid, pRM4, is finally transformed into electrocompetent cells of Xanthomonas sontii, a gram-negative phytobacterium, for final genomic integration/excision to obtain an in-frame gene deletion mutant (PPL1RM15). Gene reconstitution for complementation is carried out by electroporating the deletion mutant with the recombinant plasmid (pRM1) carrying the wild-type allele. Clean gene mutation, allele restoration, and plasmid excision are confirmed using whole-genome sequencing.
Key features
• The protocol is cost-effective and simple, eliminating the need for restriction enzymes and multiple sets of lengthy primers with restriction sites.
• The protocol requires only kinase, ligase, and polymerase, along with three sets of standard-sized desalted primers.
• The protocol is unidirectional; no need to create two different recombinant plasmids for gene deletion and complementation by reconstitution of the full-length gene.
• The difference in size between empty and recombinant vectors facilitates the easy screening of transformed Escherichia coli colonies through colony PCR.
• The strategy can be applied to any bacteria using a suitable suicide vector with appropriate positive and negative selection markers.
Keywords: Markerless gene deletion (无标记基因缺失)Graphical overview
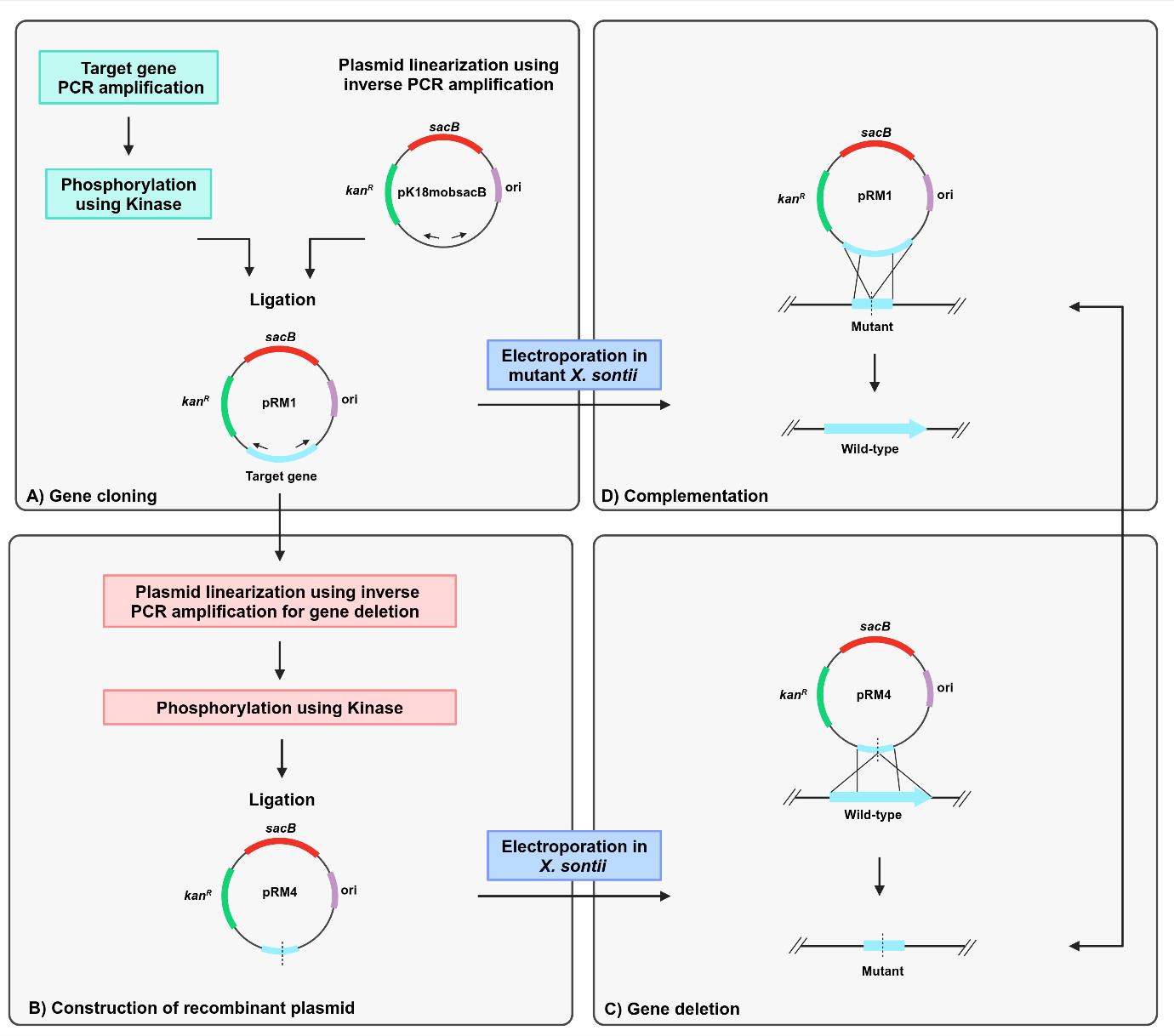
Background
Restriction enzyme–mediated cloning is complicated and requires multiple time-consuming steps. The first part is the introduction of suitable restriction sites in the target gene PCR product and the vector for cloning. Second, it is required to create distinct recombinant vectors for deletion and complementation through gene reconstitution [1]. The plasmid pK18mobsacB is a cloning vector that facilitates markerless gene deletion in gram-negative bacteria through homologous recombination [2]. The vector has a kanR marker, which confers kanamycin resistance, and a sacB gene, which confers sucrose sensitivity through the levan synthesis in the presence of sucrose [3]. A deleted version of the target gene is generated using primers with restriction sites and then cloned into pK18mobsacB, followed by allelic exchange through homologous recombination. Gene complementation is carried through reconstitution by amplification and restriction enzyme–mediated cloning of the full-length gene in pk18mobsacB and further homologous recombination in the deletion mutant.
Here, we describe a protocol for gene deletion and complementation through reconstitution in Xanthomonas sontii, a gram-negative bacterium and rice endophyte, using a pK18mobsacB-based inverse-PCR strategy without using any restriction enzyme. The standard M13 region of pK18mobsacB was used to design inverse primers. The amplified target gene was phosphorylated with kinase and ligated with the linear plasmid. Further, to generate the in-frame deletion mutant, primers facing tail-to-tail situated 300–400 bp inside from the ends of the target gene were used to amplify the recombinant plasmid, excluding the gene region to be deleted. This was followed by kinase treatment and ligation. The recombinant plasmid with the deleted gene was electroporated into the Xanthomonas cells, leading to the deletion of the target gene in the chromosome through homologous recombination. In a similar way, the recombinant plasmid with the target gene was electroporated into the gene deletion mutant of Xanthomonas sontii, leading to the restoration of the full-length target gene in the chromosome through homologous recombination.
This unidirectional method is fast, accurate, and marker-free and can be easily used to delete any gene in the chromosome and carry out chromosomal complementation through allele reconstitution independent of the reliability of restriction sites or enzymes. Further considering the average length of a gene in bacteria (i.e., 1–2 kb), the protocol is applicable for a typical gene size. Moreover, the protocol is developed in a non-model organism with high GC content. Hence, it will readily apply to organisms with much lower GC contents. This protocol is applicable in both gram-positive and gram-negative bacteria with suitable suicide vectors with negative selection markers. Counterselection markers like sacB mediating sucrose toxicity are well-suited for gram-negative bacteria and a few gram-positive bacteria like Corynebacterium and Mycobacterium [2,4]. However, for other gram-positive bacteria, the mutated version of the phenylalanine tRNA transferase (pheS*) protein [5], mazF toxin [6], or any other suitable negative selection markers can be deployed.
Materials and reagents
Biological materials
1. Xanthomonas sontii PPL1T [7]
2. Escherichia coli Top10 cells (Invitrogen, catalog number: C404010)
3. pK18mobsacB plasmid [2]
Reagents
1. Quick-DNATM Fungal/Bacterial Miniprep kit (Zymo Research, catalog number: D6005)
2. GeneJET Plasmid Miniprep kit (Thermo Scientific, catalog number: K0502)
3. 5× FIREPol® master mix (Solis BioDyne, catalog number: 04-12-00115)
4. Nuclease-free water (MP Biomedicals, catalog number: 112450204)
5. Agarose (Sigma-Aldrich, catalog number: A9539)
6. Ethidium bromide solution (Sigma-Aldrich, catalog number: E1510)
7. 6× DNA loading buffer (Real-Gene Labs, catalog number: 530001)
8. Quick-load® 100 bp DNA ladder (New England Biolabs Inc., catalog number: N0467S)
9. Quick-load® 1 kb DNA ladder (New England Biolabs Inc., catalog number: N0468S)
10. GeneJET Gel Extraction kit (Thermo Scientific, catalog number: K0691)
11. GeneJET PCR Purification kit (Thermo Scientific, catalog number: K0701)
12. Phusion DNA polymerase (Thermo Scientific, catalog number: F-530S)
13. 5× Phusion HF buffer (Thermo Scientific, catalog number: F-518)
14. dNTP mix (Fermentas, catalog number: R0192)
15. T4 Polynucleotide kinase (New England Biolabs Inc., catalog number: M0201S)
16. 10× T4 DNA ligase buffer with 10 mM ATP (New England Biolabs Inc., catalog number: B0202A)
17. T4 DNA ligase (New England Biolabs Inc., catalog number: M0202S)
18. Kanamycin sulfate (Sigma-Aldrich, catalog number: 60615)
19. 0.22 μm PVDF filter (Whatman, catalog number: 9913-2502)
20. 5 mL syringe (Dispo Van)
21. 0.025 μM desalted primers (Sigma-Aldrich)
22. DifcoTM nutrient agar (BD, catalog number: 213000)
23. DifcoTM nutrient broth (BD, catalog number: 234000)
24. DifcoTM LB broth, Miller (BD, catalog number: 244620)
25. DifcoTM LB agar, Miller (BD, catalog number: 244520)
26. Sucrose (Sigma-Aldrich, catalog number: S0389)
27. Calcium chloride (CaCl2) dihydrate (Sigma-Aldrich, catalog number: C5080-500G)
28. Glycerol (Qualigens, catalog number: Q15457)
29. Tris base (Sigma-Aldrich, catalog number: 648310-500GM)
30. Glacial acetic acid (Qualigens, catalog number: Q11007)
31. EDTA disodium dihydrate (Sisco Research Lab. Pvt. Ltd., catalog number: 43272)
Solutions
1. Nutrient agar with 5% sucrose (see Recipes)
2. 50× TAE buffer (see Recipes)
3. Kanamycin sulfate stock 50 mg/mL (see Recipes)
4. 300 mM sucrose (see Recipes)
5. 100 mM CaCl2 + 15% glycerol (see Recipes)
6. 100 mM CaCl2 (see Recipes)
Recipes
1. Nutrient agar with 5% sucrose
| Reagent | Quantity or volume |
|---|---|
| Nutrient agar | 23 g |
| Sucrose | 50 g |
| Distilled water | 1,000 mL |
Note: Sterilize media by autoclaving at 121 °C for 15 min.
2. 50× TAE buffer
| Reagent | Quantity or volume |
|---|---|
| Tris base | 242 g |
| Glacial acetic acid | 57.1 mL |
| 0.5 mM EDTA | 18.612 g |
| Distilled water | Up to 1,000 mL |
Note: 50× TAE is diluted with distilled water (1:50) to make 1× TAE for the preparation of agarose gel.
3. Kanamycin sulfate stock 50 mg/mL
| Reagent | Quantity or volume |
|---|---|
| Kanamycin sulfate | 250 mg |
| Distilled water | 5 mL |
Note: Filter sterilize the solution with a 0.22 μm syringe filter.
4. 300 mM Sucrose
| Reagent | Quantity or volume |
|---|---|
| Sucrose | 5.134 g |
| Distilled water | 50 mL |
Note: Filter sterilize the solution with a 0.22 μm syringe filter.
5. 100 mM CaCl2 + 15% glycerol
| Reagent | Quantity or volume |
|---|---|
| CaCl2·2H2O | 0.735 g |
| Glycerol | 7.5 mL |
| Distilled water | Up to 50 mL |
6. 100 mM CaCl2
| Reagent | Quantity or volume |
|---|---|
| CaCl2·2H2O | 1.47 g |
| Distilled water | 100 mL |
Equipment
1. Micropipettes (Eppendorf)
2. Nanodrop (DeNovix, catalog number: DS-11 FX)
3. UV Transilluminator (Wealtac, model: HD-25, 365 nm)
4. Gel electrophoresis assembly (Bio-Rad, model: 200/2.0)
5. Mastercycler gradient (Eppendorf, catalog number: 950000023)
6. Centrifuge (Eppendorf, model: 5810 R)
7. Microcentrifuge (Sigma, model: 1-15P)
8. Thermomixer comfort (Eppendorf, catalog number: 5355 000.011)
9. Electroporator (Bio-Rad, catalog number: 1652660)
10. Incubators at 28 °C and 37 °C (Innova®, model: Innova® 42R Inc/Ref shaker)
Software and datasets
1. AmplifX v2.0.7 [8]
2. Primer3 v0.4.0 (https://primer3.ut.ee/)
3. MEGA-X v10.2.6 [9]
4. NEBiocalculator (https://nebiocalculator.neb.com/)
Procedure
文章信息
稿件历史记录
提交日期: Jan 2, 2025
接收日期: Apr 15, 2025
在线发布日期: May 6, 2025
出版日期: May 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Rana, R., Sharma, A., Dutta, A. and Patil, P. B. (2025). I-PREFR: Inverse PCR-Based Restriction Enzyme FRee Unidirectional Strategy for Rapid Markerless Chromosomal Gene Deletion and Reconstitution in Bacteria Using Suicide Vectors. Bio-protocol 15(10): e5314. DOI: 10.21769/BioProtoc.5314.
分类
微生物学 > 微生物遗传学 > 基因组编辑
分子生物学 > DNA > 染色体工程
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













