- EN - English
- CN - 中文
RNA PolyA Tailing Assay to Qualitatively Analyze Circular RNA Manufacturing
RNA加尾实验用于环状RNA合成的质量分析
发布: 2025年05月20日第15卷第10期 DOI: 10.21769/BioProtoc.5310 浏览次数: 1660
评审: Joyce ChiuAnonymous reviewer(s)

相关实验方案

用于Poly(A)纯化mRNA中N6-甲基腺苷定量的改良型m6A-ELISA方法
Wei Yee Chan [...] Folkert J. Van Werven
2025年06月20日 3006 阅读
Abstract
A covalently closed loop structure provides circular RNA (circRNA) with more stability than conventional RNAs in linear form, making circRNA an emerging tool in RNA therapeutics. The qualification and quantification of circRNA after production is critical for its design and effectiveness assessments, particularly when the following applications could be affected by byproduct RNAs. Despite PCR-based methods effectively detecting low-abundance circRNA, they are unsuitable for assessing uncircularized RNA in a mass production fraction to maintain quality control. Here, we present a straightforward protocol for evaluating uncircularized byproduct RNAs from circRNA production. This method enrolls the template-independent RNA polymerase activity to add adenine tails (polyA) to the 3' ends of a linear RNA, making it easy to distinguish trace byproducts or uncircularized RNA from a pool of mass circRNA products. With conventional linear RNA and RNase R-treated circRNA as the positive and negative controls, the purity of a circRNA preparation could be readily resolved. Regardless of circRNA production strategies, this protocol provides a reliable and practical way to ensure the consistent quality of homemade circRNAs or to recheck circRNA quality from commercial manufacturing.
Key features
• The RNA circularization assessment relies on detecting a free 3' end OH group of the non-circRNA in the circRNA products.
• An RNA denaturing electrophoresis is required to detect nucleotide length changes of the non-circRNA post-3' end tailing.
• Rather than quantitatively, the molecular weight changes qualitatively highlight the trace non-circRNA in a circRNA preparation.
• While there are different techniques for producing circRNA, the tailing method is effective for most, detecting leftover linear RNA contaminants.
Keywords: RNA tailing/telling assay (RNA加尾/检测实验)Graphical overview
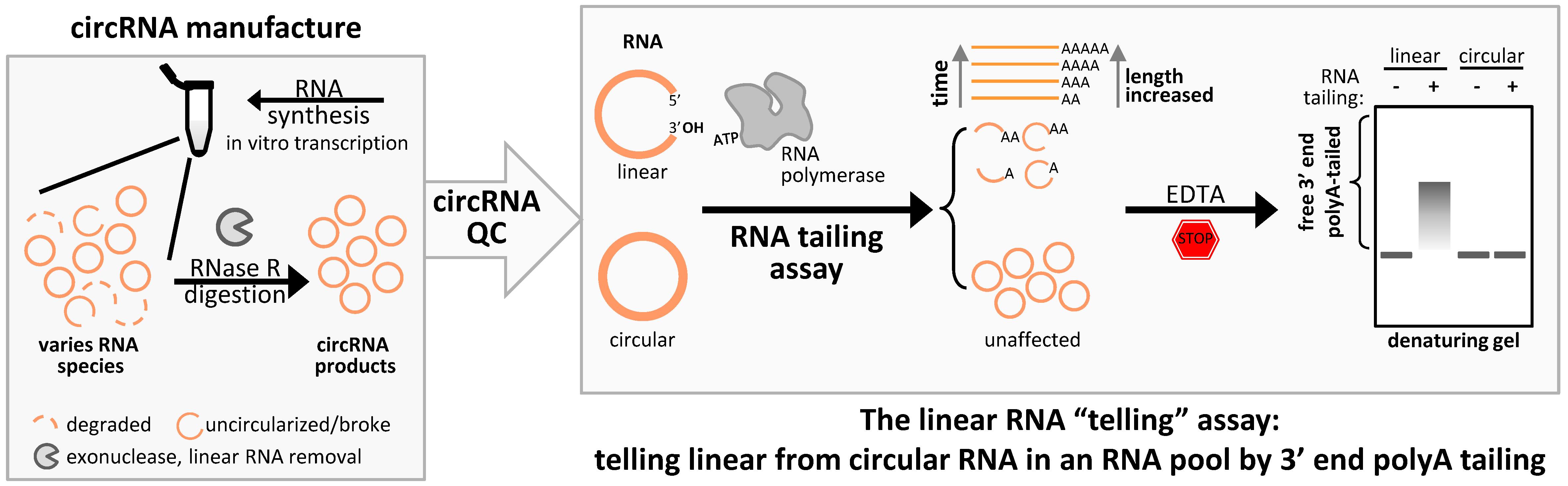
Background
Circular RNAs (circRNAs) are unique RNAs characterized by their covalently closed-loop structures. These structures confer increased stability compared to their linear counterparts by rendering them resistant to exonuclease-mediated degradation [1]. This natural stability makes circRNAs promising candidates for RNA-based therapeutics, including gene regulation, biomarker development, and targeted drug delivery [2]. As the diversity of circRNA design strategies and applications continues to expand, accurately assessing circRNA yield and purity has become an emerging and critical need.
Current circRNA detection methods [3] mainly detect and quantify a relatively small amount of circRNAs in a specific context. High-throughput RNA sequencing (RNA-Seq), combined with bioinformatics, analyzes various types of genome-wide circRNAs present in low abundance [4]. By targeting the junction sequences of RNA back-splicing/ligation, quantitative reverse transcription PCR (qRT-PCR) offers sensitive and specific quantification for trace circRNA from an RNA pool [5]. Even though these approaches effectively detect circRNA, none directly assess the accompanying linear RNAs in the same pool. This is especially crucial for quality control and downstream applications of circRNA after mass production.
Linear RNA contaminants can be found in a circRNA preparation for several reasons, such as yet-to-be circularized RNA, somehow disconnected circRNAs, or any byproduct RNAs with free 3' ends in circRNA production processes. Here, we present a streamlining assay that leverages the activity of template-independent RNA polymerase to append adenine tails (polyA) to the 3' ends of linear RNAs [6]. As a consequence, the prerequisite for the success of this polyA tailing assay significantly relies on optimizing denaturing formaldehyde gel conditions and testing appropriate tailing assay parameters in-house. Denaturing gel electrophoresis offers better resolution and prevents RNA degradation, reducing misleading band shifts compared to non-denaturing TBE gels. This strategy clearly tells that any linear RNA remains in the circRNA products, allowing for a reliable assessment of circRNA purity, predominantly when the sample is mainly composed of circRNA.
Exonuclease resistance is a circRNA signature, distinguishing it from linear RNA [7]. However, RNase R can be used for linear RNA removal in circRNA preparation rather than for detecting circRNA. To evaluate linear RNA in the bulk of circRNA preparation more straightforwardly and intuitively, we repurposed the polyA tailing assay from its primary application for in vitro mRNA preparation [8]. This method provides an effective and practical approach to assessing circRNA integrity.
Materials and reagents
Reagents
1. Agarose biomax (Hispanagar, CAS number: 9012-36-6)
2. Diethyl pyrocarbonate (DEPC) (VWR, catalog number: E174-25G)
3. E. coli PolyA polymerase (NEB, catalog number: M0276S)
4. EDTA, disodium salt, dihydrate, crystal (J. T. Baker, catalog number: 8993-01)
5. Ethyl alcohol, 99%, HPLC/ACS (Everdine Technology, CAS number: 64-17-5)
6. Formaldehyde solution (Sigma, catalog number: F1635)
7. GelRed nucleic acid gel stain (Biotium, catalog number: 41003)
8. MEGAclear Transcription Clean-Up kit (Invitrogen, catalog number: AM1908)
9. MOPS buffer (10×) (ZGENEBIO, catalog number: GB33)
10. RNA loading dye (2×) (NEB, catalog number: B0363S)
11. RNase R (Abcam, catalog number: ab286929)
12. ssRNA ladder (NEB, catalog number: N0362S)
13. TBE buffer (5×) (VWR, catalog number: J885-5L)
14. Adenosine 5'-triphosphate (ATP) (NEB, catalog number: P0756S)
Solutions
1. DEPC-treated water (see Recipes)
2. MOPS buffer (1×) (see Recipes)
3. Denaturing formaldehyde agarose gel (1%) (see Recipes)
4. TBE buffer (0.5×) (see Recipes)
5. Non-denaturing TBE agarose gel (1%) (see Recipes)
6. EDTA solution (0.5 M) (see Recipes)
7. RNA polyA tailing reaction mixture (see Recipes)
8. EDTA stop solution (see Recipes)
9. RNase R digestion (see Recipes)
Recipes
1. DEPC-treated water
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DEPC | 0.1% | 1 mL |
| Deionized water | 999 mL | |
| Total | 1 L |
Stir overnight. To remove DEPC, autoclave the solution for 15–45 min at 15 psi.
2. MOPS buffer (1×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MOPS buffer (10×) | 1× | 50 mL |
| DEPC-treated water | 450 mL | |
| Total | 500 mL |
3. Denaturing formaldehyde agarose gel (1%)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1% | 0.5 g |
| MOPS buffer (1×) | 42 mL | |
| Formaldehyde | 2 M | 8 mL |
| Total | 50 mL |
4. TBE buffer (0.5×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| TBE buffer (5×) | 0.5× | 100 mL |
| DEPC-treated water | 900 mL | |
| Total | 1 L |
5. Non-denaturing TBE agarose gel (1%)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1% | 0.5 g |
| TBE (1×) | 50 mL | |
| GelRed nucleic acid gel stain (10,000×) | 1× | 5 μL |
| Total | 50 mL |
6. EDTA solution (0.5 M)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EDTA | 0.5 M | 18.61 g |
| Deionized water | 80 mL | |
| DEPC | 0.1% | 100 μL |
| Total | 100 mL |
Adjust the final volume to 100 mL using deionized water and stir overnight. To remove DEPC, autoclave the solution for 15–45 min at 15 psi.
7. RNA polyA tailing reaction mixture
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA sample | 1.25 μg | 15 μL |
| E. coli PolyA polymerase reaction buffer (10×) | 1× | 2 μL |
| ATP (10 mM) | 1 mM | 2 μL |
| E. coli PolyA Polymerase | 5 U | 1 μL |
| Total | 20 μL |
8. EDTA stop solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA polyA tailing reaction mixture | 20 μL | |
| EDTA (50 mM) | 10 mM | 5 μL |
| Total | 25 μL |
9. RNase R digestion
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| RNA sample | 100 μg | 88 μL |
| RNase R reaction buffer (10×) | 1× | 10 μL |
| RNase R | 20 U | 2 μL |
| Total | 100 μL |
Adjust the final volume to 100 μL using DEPC-treated water.
Laboratory supplies
1. Eppendorf DNA LoBind tubes (Eppendorf, catalog number: 022431021)
2. 0.1–10 μL sterilized filter tips (GILSON, catalog number: F171203)
3. 10–100 μL sterilized filter tips (GILSON, catalog number: F171403)
4. 2–200 μL sterilized filter tips (GILSON, catalog number: F171503)
5. 50–1,200 μL sterilized filter tips (GILSON, catalog number: F171803)
Equipment
1. 37 °C incubator (Astec, catalog number: SCA-165DS)
2. Block incubator (Astec, catalog number: BI-525)
3. Gel electrophoresis system (Major Science, catalog number: MJ-105-S)
4. Electrophoresis power supply (Thermo, catalog number: EC250-90)
5. Orbital shaker (TKS, catalog number: OS701)
6. Microcentrifuge (Eppendorf, catalog number: 5424)
Software and datasets
1. Quantum CX5 Gel Imaging System (VILBER, BIO VISION)
2. ImageJ (NIH, 1.54p)
Procedure
文章信息
稿件历史记录
提交日期: Mar 10, 2025
接收日期: Apr 16, 2025
在线发布日期: May 5, 2025
出版日期: May 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Chuang, Z., Su, C., Wang, H. and Yu, C. (2025). RNA PolyA Tailing Assay to Qualitatively Analyze Circular RNA Manufacturing. Bio-protocol 15(10): e5310. DOI: 10.21769/BioProtoc.5310.
分类
分子生物学 > RNA > RNA 修饰
生物化学 > RNA > RNA结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











