- EN - English
- CN - 中文
Mapping the Simultaneously Accessible and ssDNA-Containing Genome With KAS-ATAC Sequencing
利用KAS-ATAC测序绘制同时具有开放性和单链DNA特征的基因组图谱
发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5306 浏览次数: 1758
评审: Laxmi Narayan MishraAnonymous reviewer(s)

相关实验方案

优化高分子量 DNA 提取方法以用于 Magnaporthaceae 及其他禾本科根部真菌的长读长全基因组测序
Michelle J. Grey [...] Mark McMullan
2025年03月20日 3268 阅读
Abstract
The KAS-ATAC assay provides a method to capture genomic DNA fragments that are simultaneously physically accessible and contain single-stranded DNA (ssDNA) bubbles. These are characteristic features of two of the key processes involved in regulating and expressing genes—on one hand, the activity of cis-regulatory elements (cREs), which are typically devoid of nucleosomes when active and occupied by transcription factors, and on the other, the association of RNA polymerases with DNA, which results in the presence of ssDNA structures. Here, we present a detailed protocol for carrying out KAS-ATAC as well as basic processing of KAS-ATAC datasets and discuss the key considerations for its successful application.
Key features
• Allows mapping of simultaneously accessible and ssDNA-containing DNA fragments.
• Describes the execution of N3-kethoxal labeling and transposition of native chromatin.
• Describes the pulldown of biotin-labeled DNA fragments and library generation.
• Describes basic KAS-ATAC data processing steps.
Keywords: KAS-seq (KAS-seq)Graphical overview
Background
In most eukaryotes, the major steps in the process of gene expression are the act of transcription itself and its activation/repression by the combined action of transcription factors (TFs) on cis-regulatory elements (cREs), i.e., promoters, enhancers, and insulators. The action of cREs can be highly complex, e.g., the input of on average ≥10 individual cREs per gene is integrated into the regulation of promoter activity in mammalian genomes [1,2]. Therefore, mapping the location and activation status of cREs genome-wide has been a key task for the comprehensive charting of regulatory networks.
To this end, a key property of active cREs—that they tend to be devoid of nucleosomes [3–5]—has been the basis for technological development, due to the fact that the absence of nucleosomal protection renders active cREs accessible to enzymatic action, whether cleavage (e.g., by DNase I [6–9]) or modification (e.g., by methyltransferases [10–12]). The enzyme with the most utility for mapping open chromatin genome-wide has turned out to be a hyperactive version of the Tn5 transposase [13,14], which allows both the labeling of accessible regions in the genome and their tagging with readily PCR-amplifiable sequencing adapters, thus greatly simplifying experimental protocols and minimizing input amount requirements, in the form of the ATAC-seq assay [14] (assay for transposase-accessible chromatin using sequencing) and its many variations.
ATAC-seq datasets also contain highly useful finer-grained information in addition to the mere location of open chromatin regions. They can be used to map nucleosome positioning around cREs [15] as well as the footprints of individual TFs associated with DNA, which also protect DNA from cleavage, even if to a lesser extent than nucleosomes [14].
The integrated action of cREs results in the modulation of transcriptional activity at promoters, with enhancers themselves also being transcribed [16]. Thus, mapping active transcription (as opposed to steady-state RNA levels, which are the result of both active transcription and the subsequent effects of intrinsic RNA stability and post-transcriptional regulation) has been another key tool for understanding the regulatory genome. To this end, multiple adaptations of nuclear run-on techniques, such as GRO-seq [17], PRO-seq [18], and others [19], have been developed. However, these are generally fairly complex protocols, and as they measure RNA rather than DNA molecules, they do not allow the simultaneous recording of both transcriptional activity and the physical state of the genome. A more recent alternative tool that does resolve these issues is the KAS-seq [20] assay (kethoxal-assisted ssDNA sequencing), based on the highly specific covalent labeling of unpaired guanine residues by N3-kethoxal. N3-kethoxal adducts can then be subjected to click chemistry–mediated biotinylation, and the DNA fragments containing ssDNA can be specifically enriched and amplified. As most ssDNA in the genome is the result of RNA polymerase bubbles, both elongating and paused (with the rest coming from active replication intermediates and some secondary structures), KAS-seq is a convenient tool for mapping polymerase engagement with the genome.
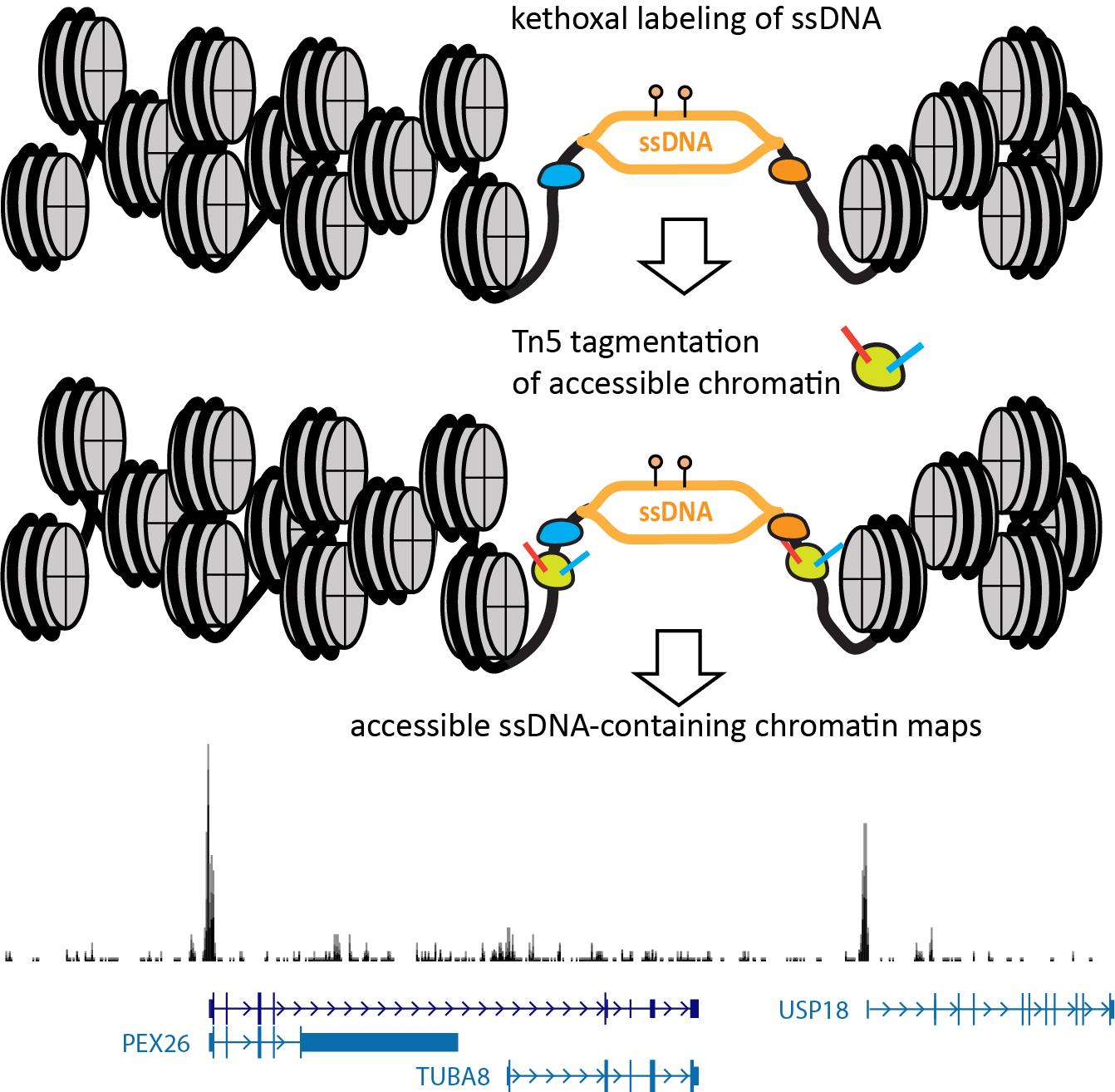
The nature of the KAS method—specifically, the fact that it labels ssDNA with permanent covalent tags—allows its extension to single-molecule multiomics readouts that capture additional modalities on the same genomic fragments. The KAS-ATAC method that we recently developed [21] enables the mapping of genomic fragments that are both physically accessible and contain ssDNA, together with nucleosome positions and TF footprints within them and their vicinity, by combining the ATAC-seq and KAS-seq assays into one (Figure 1). This is accomplished by first quickly incubating live cells with N3-kethoxal, then washing the kethoxal away, and proceeding immediately to the native chromatin transposition step of the ATAC-seq protocol. The resulting Tn5-tagged accessible DNA fragments are purified, kethoxal-labeled guanines are biotinylated via a click chemistry reaction, then ssDNA-containing fragments are specifically enriched using a streptavidin pull-down. Finally, PCR amplification is carried out on beads to generate final Illumina-compatible libraries.
This protocol describes the step-by-step execution of the KAS-ATAC assay and the basic computational processing of the resulting datasets.
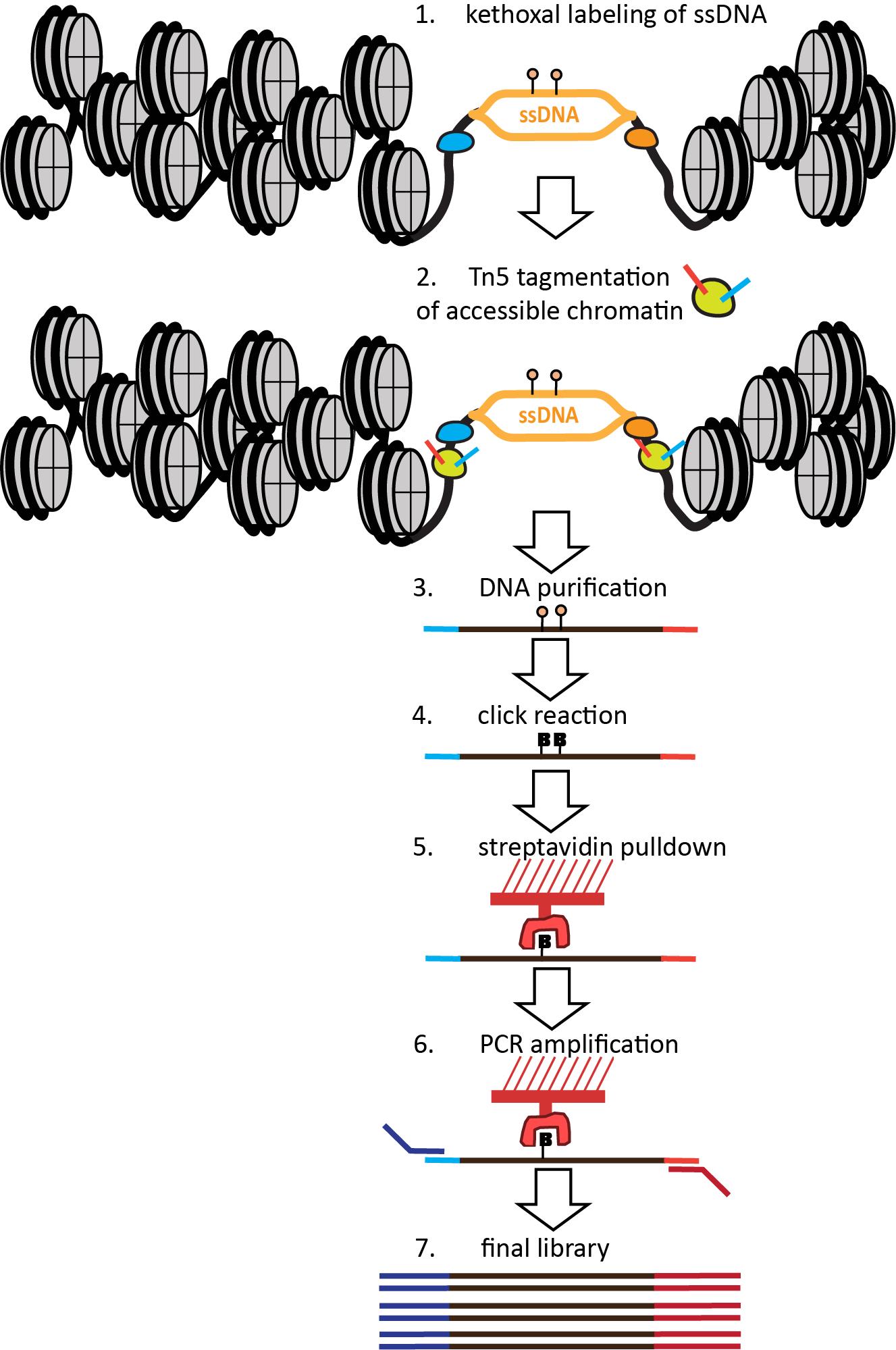
Figure 1. Outline of the KAS-ATAC assay. First, ssDNA is labeled using a quick treatment with N3-kethoxal. Next, the kethoxal is washed away, nuclei are isolated, and native transposition is carried out to label accessible chromatin. DNA is then isolated and subjected to a click reaction to attach biotin to kethoxal adducts. Biotin-labeled DNA fragments are then specifically pulled down using streptavidin and PCR-amplified on beads. Final libraries are sequenced on an Illumina instrument.
Materials and reagents
Reagents
1. Tn5 transposase; can be obtained from the Nextera XT DNA Library Preparation Kit offered by Illumina (catalog number: FC-131-1024), separately from Illumina (catalog number: 20034197 or 20034198), from Diagenode (catalog number: C01070012-30), and also from several other commercial vendors. It can also be made locally following previously published protocols [13], which is the most cost-effective approach, especially for large-scale projects. The oligo sequences needed for the transposome assembly are the following:
Tn5MErev: /5Phos/CTGTCTCTTATACACATCT
Tn5ME-A: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG
Tn5ME-B: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG
If homemade Tn5 is used, its activity should be carefully characterized relative to standard enzymatic formulations before production-scale use (using ATAC-seq enrichment metrics as a criterion).
2. Sequencing primers/adapters: PCR and indexing primers/adapters can be obtained from the Nextera XT DNA Library Preparation Kit or equivalent kits. Alternatively, custom-designed and synthesized oligos can also be used.
The i7 primer sequence is: 5'-CAAGCAGAAGACGGCATACGAGAT[i7]GTCTCGTGGGCTCGG-3'
The i5 sequence is: 5'AATGATACGGCGACCACCGAGATCTACA
C[i5]TCGTCGGCAGCGTC-3’
Where [i7] and [i5] are the index sequences (typically 8-bp long). Typical [i7] and [i5] index sequences are the following:
For [i7]:
701. TAAGGCGA
702. CGTACTAG
703. AGGCAGAA
704. TCCTGAGC
705. GGACTCCT
706. TAGGCATG
707. CTCTCTAC
708. CAGAGAGG
709. GCTACGCT
710. CGAGGCTG
711. AAGAGGCA
712. GTAGAGGA
713. GTCGTGAT
714. ACCACTGT
715. TGGATCTG
716. CCGTTTGT
717. TGCTGGGT
718. GAGGGGTT
719. AGGTTGGG
720. GTGTGGTG
721. TGGGTTTC
722. TGGTCACA
723. TTGACCCT
724. CCACTCCT
For [i5]:
501. TAGATCGC
502. CTCTCTAT
503. TATCCTCT
504. AGAGTAGA
505. GTAAGGAG
506. ACTGCATA
507. AAGGAGTA
508. CTAAGCCT
Dissolve and dilute to 25 μM.
1. N3-kethoxal (ApexBio, catalog number: A8793)
2. Dimethyl formamide (Sigma, catalog number: 227056-100ML)
3. 1× PBS buffer solution pH 7.4 (Thermo Fisher Scientific, catalog number: 10010031)
4. 10× PBS buffer solution pH 7.4 (Thermo Fisher Scientific, catalog number: 70011044)
5. 1 M Tris-HCl pH 7.4 (Thermo Fisher Scientific, catalog number: J60202.K2)
6. 5 M NaCl (Thermo Fisher Scientific, catalog number: A57006)
7. 1 M MgCl2 (Thermo Fisher Scientific, catalog number: J61014.AK)
8. IGEPAL CA-630 detergent (Sigma, catalog number: 11332465001; supplied as a 10% solution)
9. Tween-20 detergent (Sigma, catalog number: 11332465001; supplied as a 10% solution); store at 4 °C
10. Digitonin detergent (Promega, catalog number: G9441; supplied as a 2% solution in DMSO); store at -20 °C
11. K3BO3 (Sigma, catalog number: B6768); prepare a carefully pH-adjusted pH 7.0 250 mM solution
12. 0.5 M EDTA, pH 8.0 (Thermo Fisher Scientific, catalog number: 15575020)
13. DBCO-PEG4-biotin (Sigma, catalog number: 760749); dissolve in DMSO to 20 mM
14. Dynabeads MyOne Streptavidin T1 beads (Thermo Fisher Scientific, catalog number: 65601)
15. NEBNext High-Fidelity 2× PCR Master Mix (NEB, catalog number: M0541S)
16. QuBit 1× dsDNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q33231)
17. MinElute PCR Purification kit (Qiagen, catalog number: 28004/28006), Zymo DNA Clean and Concentrator kit (Zymo, catalog number: D4013/D4014), or equivalent
18. Nuclease-free H2O
Solutions
1. ATAC-RSB buffer (master stock) (see Recipes)
2. ATAC-RSB-lysis buffer (see Recipes)
3. Lysis wash buffer (ATAC-RSB-wash) (see Recipes)
4. 2× TD buffer (see Recipes)
5. Tween washing buffer (TWB) (see Recipes)
6. 2× binding buffer (see Recipes)
7. Transposition mix (see Recipes)
Recipes
1. ATAC-RSB buffer (master stock)
10 mM Tris-HCl pH 7.4
10 mM NaCl
3 mM MgCl2
Store at 4 °C.
2. ATAC-RSB-lysis buffer
10 mM Tris-HCl pH 7.4
10 mM NaCl
3 mM MgCl2
0.1% IGEPAL CA-630
0.1% Tween-20
0.01% Digitonin
Prepare immediately before use.
3. Lysis wash buffer (ATAC-RSB-wash)
10 mM Tris-HCl pH 7.4
10 mM NaCl
3 mM MgCl2
0.1% Tween-20
Prepare immediately before use.
4. 2× TD buffer
20 mM Tris-HCl pH 7.6
10 mM MgCl2
20% dimethyl formamide
Store at -20 °C.
It can also be obtained commercially, i.e., from Illumina (catalog number: 20034197/20034198) or Diagenode (catalog number: C01019043).
5. 1× tween washing buffer (TWB)
5 mM Tris-HCl pH 7.4
0.5 mM EDTA
1 M NaCl
0.05% Tween 20
Store at 4 °C.
6. 2× binding buffer
10 mM Tris-HCl pH 7.4
1 mM EDTA
2 M NaCl
Store at room temperature (RT).
7. Transposition mix (50 μL volume)
25 μL of 2× TD buffer
22.5 μL of H2O
2.5 μL of Tn5
Prepare immediately before use.
Laboratory supplies
1. 1.5-mL microcentrifuge tubes, preferably low protein and DNA binding (e.g., Eppendorf, catalog number: EP022431021)
2. 15 and 50 mL tubes
3. QuBit-compatible 200 μL PCR tubes/strips
4. TapeStation D1000 or HS D1000 tape and reagents (Agilent) or equivalent, e.g., BioAnalyzer (Agilent)
5. Pipette tips (1,000, 200, and 20 μL)
6. Micropipettes
Equipment
1. Incubator (37 °C) or thermomixer
2. Tabletop centrifuge
3. Thermal cycler
4. Qubit fluorometer or equivalent
5. TapeStation (Agilent) or equivalent, e.g., BioAnalyzer (Agilent)
6. Magnetic stand for 1.5 mL tubes
7. Magnetic stand for 200 μL PCR tubes/strips
8. Rotator for 1.5 mL tubes
Software and datasets
1. Bowtie [22] (http://bowtie-bio.sourceforge.net/index.shtml) or Bowtie2 [23] (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
2. samtools [24]: http://www.htslib.org/
3. PicardTools https://broadinstitute.github.io/picard/
4. UCSC Genome Browser [25, 26] utilities: http://hgdownload.cse.ucsc.edu/admin/exe/
5. Python (version 2.7 or higher): https://www.python.org/
6. Custom Python scripts (https://github.com/georgimarinov/GeorgiScripts)
Procedure
文章信息
稿件历史记录
提交日期: Feb 1, 2025
接收日期: Apr 9, 2025
在线发布日期: Apr 24, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Marinov, G. K. and Greenleaf, W. J. (2025). Mapping the Simultaneously Accessible and ssDNA-Containing Genome With KAS-ATAC Sequencing. Bio-protocol 15(9): e5306. DOI: 10.21769/BioProtoc.5306.
分类
生物信息学与计算生物学
系统生物学 > 基因组学 > 测序
分子生物学 > DNA > 染色质可接近性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











