- EN - English
- CN - 中文
Reconstruction of Single-Neuron Projectomes in Mice
小鼠单神经元投射图谱的重建
(*contributed equally to this work) 发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5300 浏览次数: 1480
评审: Geoffrey C. Y. LauShivaprasad H. Sathyanarayana
Abstract
Reconstructing single-neuron projectomes is essential for mapping the mesoscopic connectome and eventually for understanding brain-wide connectivity and diverse brain functions. The combination of sparse labeling techniques and large-scale and high-resolution optical imaging technologies has been revolutionizing the brain-wide reconstruction of single-neuron morphologies, as exemplified by the dataset for over 10,100 single-neuron projectomes of hippocampal neurons. Here, we illustrate a comprehensive protocol for large-scale single-neuron reconstruction in the mouse brain. This includes key steps and examples in imaging data preprocessing, neurite tracing, and registration into a template brain. These procedures enable efficient and accurate large-scale morphological reconstruction of single neurons in the mouse brain.
Key features
• Rigorous and effective single-neuron reconstruction from raw imaging data.
• Multi-person tracing and quality control for the accuracy of single-neuron tracing results.
• Precise image registration based on landmark drawings of selected brain regions.
Keywords: Single-neuron reconstruction (单神经元重建)Graphical overview
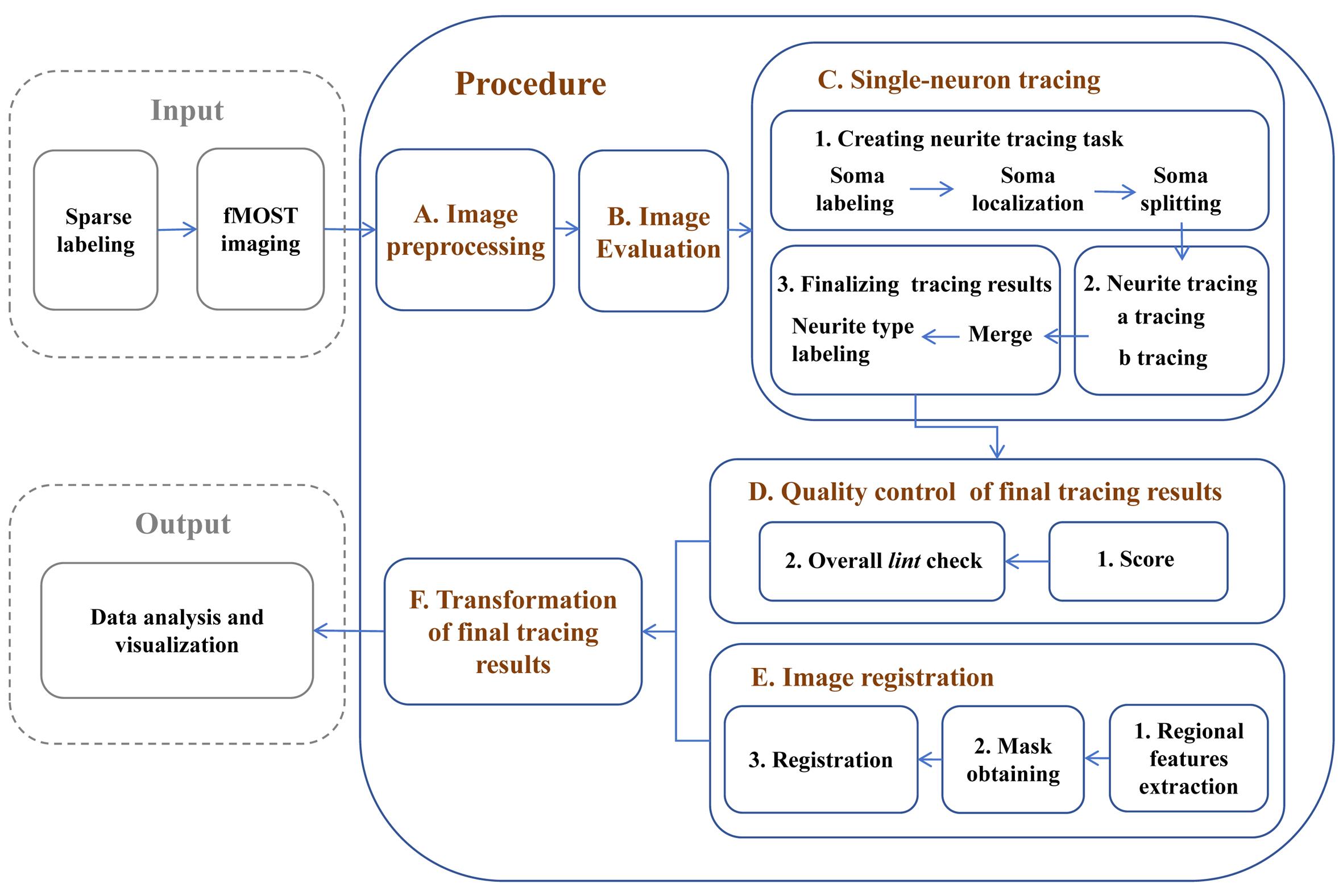
Reconstruction of single-neuron projectomes in mice
Background
Reconstructing the morphology of neurons at the single-cell level provides new insights into various neuroscience issues, including neural cell classification, elucidation of information transmission among different neuron types, and clarification of the potential functions of distinct brain regions [1–3]. Due to the diversity and complexity of neuron morphology, establishing an efficient procedure for single-neuron reconstruction that ensures both the integrity and accuracy of the results is essential for obtaining large-scale single-neuron reconstruction data and conducting subsequent analysis [4]. In recent years, advancements in sparse labeling techniques [5,6] and advanced imaging systems, such as fluorescence micro-optical sectioning tomography (fMOST) [7], have enabled the acquisition of continuous section imaging results for labeled neurons. However, there is still a need for a systematic approach and optimization strategies for reconstructing single-neuron morphologies across the entire brain from raw imaging data. In this protocol, we first preprocess the fMOST imaging results and establish quality control standards to assist researchers in selecting high-quality imaging samples suitable for continuous tracing of single-neuron morphologies across the entire brain. Next, we developed a neuron soma localization tool that allows for selective tracing within the region of interest, mitigating interference from labeled neurons in other regions—an ongoing challenge in dense labeling approaches [8,9]. For the successfully traced neurons, by using the semi-automated tracing software Fast Neurite Tracer (FNT) [10], we significantly enhanced the accuracy of the tracing results by increasing quality control steps. Additionally, we distinguished labeled axonal and dendritic structures, enabling the analysis of morphological relationships among various neuronal structures. As a final step in our reconstruction pipeline, we enhanced the spatial accuracy of the reconstructed neurons by implementing a registration method that utilizes features extracted from multiple brain subregions. The single-neuron reconstruction results obtained through these methods can be directly applied to data visualization and analysis. This procedure has been utilized in studies of single-neuron projectomes in multiple mouse brain regions, building the currently largest dataset of whole-brain projectome maps for 10,100 single neurons in the mouse hippocampus [11,12]. It provides a foundation for future cross-species brain mapping research, including non-human primate studies, and offers guidance for research on brain diseases and brain-inspired artificial intelligence.
Equipment
1. Workstation PC [e.g., Dell, model: Intel(R) Xeon(R) W-2135 CPU @ 3.70GHz, Windows 10 Pro 64, 64 GB RAM]
2. High-performance computing cluster
Software and datasets
Software
1. Fast Neurite Tracer, https://zenodo.org/record/5981001
2. Fiji, https://fiji.sc
3. Amira, https://www.thermofisher.com/, commercial license required
4. Advanced Normalization Tools (ANTs), https://docs.itk.org/en/latest/download.html
Tools and Codes
1. All tools and codes have been deposited to GitHub: https://github.com/BiyuRen/Single_Neuron_Reconstruction
Procedure
文章信息
稿件历史记录
提交日期: Dec 26, 2024
接收日期: Apr 3, 2025
在线发布日期: Apr 18, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ren, B., Shi, X., Zhao, B., Xu, C. and Wang, X. (2025). Reconstruction of Single-Neuron Projectomes in Mice. Bio-protocol 15(9): e5300. DOI: 10.21769/BioProtoc.5300.
- Qiu, S., Hu, Y., Huang, Y., Gao, T., Wang, X., Wang, D., Ren, B., Shi, X., Chen, Y., Wang, X., et al. (2024). Whole-brain spatial organization of hippocampal single-neuron projectomes. Science. 383(6682): eadj9198. https://doi.org/10.1126/science.adj9198
分类
生物信息学与计算生物学
神经科学 > 神经解剖学和神经环路 > 脑神经
神经科学 > 基础技术 > 组织解剖
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













