- EN - English
- CN - 中文
Optimized Protocol for DNA Extraction in Three Theobroma Species
三种可可属植物的DNA提取优化方法
发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5297 浏览次数: 2075
评审: Anonymous reviewer(s)
Abstract
DNA extraction is a crucial step in molecular biology research, particularly for genetic and genomic analyses. These studies require a high concentration of high-quality DNA, which is often a challenge for underexplored species or when the available plant material consists of aged tissue. To address these challenges, the cetyltrimethylammonium bromide (CTAB)-based DNA extraction method has been optimized to improve efficiency and yield. The process begins with an overnight incubation of plant tissue macerated with liquid nitrogen in a solution containing a high concentration of CTAB (4%). Subsequently, the mixture undergoes two washes with chloroform: isoamyl alcohol. The nucleic acids are then precipitated using isopropanol, followed by a wash with 70% ethanol to ensure purity. Finally, the purified DNA is resuspended in ultrapure water. This optimized procedure produces high-quality DNA suitable for various downstream applications, including PCR and sequencing, even from older leaves of the three Theobroma species: T. cacao, T. bicolor, and T. grandiflorum. Additionally, this protocol significantly enhances throughput and allows for the parallel processing of a substantially larger number of samples compared to conventional techniques.
Key features
• An efficient CTAB-based DNA extraction protocol provides high-quality nucleic acids from older leaves by increasing CTAB concentration and incubation time in lysis buffer.
• Provides reliable yields in the three Theobroma species: T. cacao, T. bicolor, and T. grandiflorum.
• A high-throughput workflow reduces processing time and increases daily sample capacity, supporting large-scale genomic investigations.
Keywords: Theobroma (可可属)Graphical overview
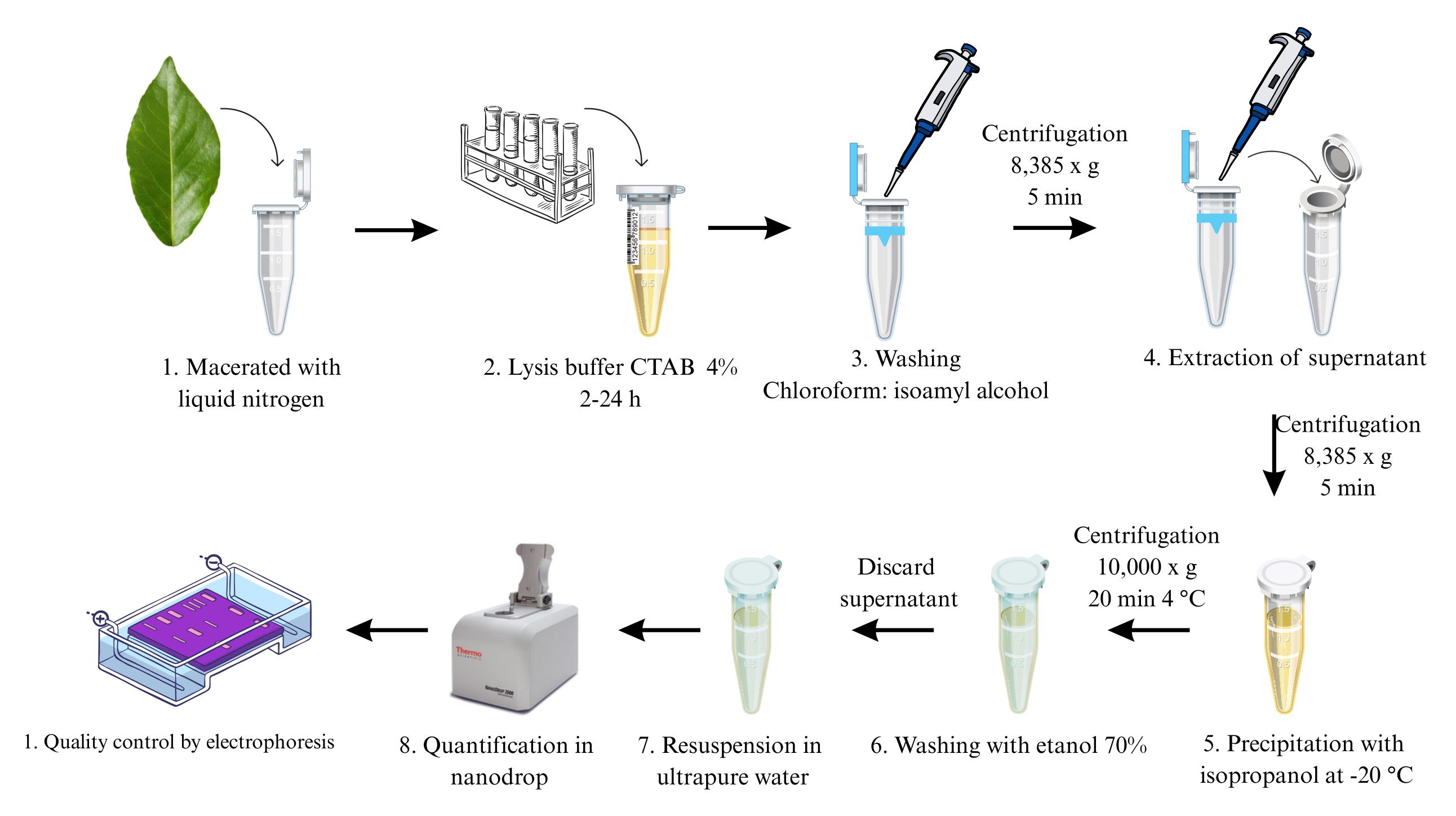
Extraction, quantification, and assessment of DNA integrity
Background
The extraction of high-quality DNA is a fundamental step in molecular biology and genomics. DNA isolation often requires high levels of concentration, purity, and integrity, particularly for library preparation and sequencing applications [1]. Despite significant advancements in DNA extraction protocols, obtaining high‐molecular‐weight DNA from challenging plant materials, such as older leaves, remains problematic [2], especially when dealing with underexplored species like T. grandiflorum and T. bicolor. Therefore, there is a need to evaluate protocols capable of extracting high-quality DNA from Theobroma species, which have promising applications. Furthermore, obtaining young tissue for DNA extraction is often impractical, particularly in native Theobroma species, which are frequently decades old and grow to considerable heights [3]. Traditional methodologies, such as the cetyltrimethylammonium bromide (CTAB)-based DNA extraction protocol, are widely used due to their adaptability and efficiency in isolating plant DNA. However, conventional CTAB protocols often yield suboptimal results when applied to samples with high levels of contaminants or structural complexity [4]. These limitations underscore the need for protocol modifications to improve DNA yield and purity, especially in underexplored species such as T. bicolor and T. grandiflorum, as well as in economically significant species like T. cacao. The protocol described here addresses these challenges by increasing the CTAB concentration in the lysis buffer and extending the incubation period. These adjustments enhance cell wall disruption and facilitate the removal of secondary metabolites, resulting in higher DNA yield and quality [5]. Additionally, the optimized protocol significantly enhances throughput, enabling the processing of a greater number of samples compared to conventional techniques. This makes it particularly suitable for large-scale genomic studies, such as comparative genomics and population genetics in cacao-related species. Beyond its use in Theobroma species, this protocol may also be adapted for other plant species with similar extraction challenges. By providing a more efficient and reliable method for isolating high-quality DNA, this protocol facilitates the exploration of genetic diversity and supports a range of molecular and genomic investigations.
Materials and reagents
Biological materials
1. Leaves of T. grandiflorum
2. Leaves of T. cacao
3. Leaves of T. bicolor
Reagents
1. Liquid nitrogen
2. β-mercapto-ethanol (BME) (Thermo Scientific, CAS: 60-24-2)
3. Chloroform (Merck, CAS: 67-66-3)
4. Isoamyl alcohol (CARLO ERBA, CAS: 123-51-3)
5. Isopropanol, AR purity (Honeywell, CAS: 67-63-0)
6. 70% ethanol, AR purity (Sigma-Aldrich, CAS: 64-17-5)
7. Ultrapure water
8. CTAB (Sigma-Aldrich, CAS: 57-09-0)
9. Sodium chloride (CARLO ERBA, CAS: 7647-14-5)
10. Ethylenediaminetetraacetic acid (EDTA) (PanReac-ITW Reagents, CAS: 6381-92-6)
11. Tris(hydroxymethyl)aminomethane (TRIS) (Thermo Fisher Scientific, CAS: 76-86-1)
12. CTAB 2%, 3%, 4% (Sigma-Aldrich, CAS: 57-09-0)
13. Proteinase K (Canvax, CAS: 39450-01-6)
14. Boric acid (Scharlau, CAS: 10043-35-3)
15. Ethidium bromide (Thermo Scientific, CAS: 1239-45-8)
16. Agarose (AMRESCO, LLC, CAS: 90-12-36-6)
17. HyperLadder 1 kb (Bioline, catalog number: BIO-33025)
18. Ethanol (Merck, catalog number: 100983)
Solutions
1. Lysis buffer CTAB 4% m/v (see Recipes)
2. CIA (see Recipes)
Recipes
1. Lysis buffer CTAB 4% m/v
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CTAB | 4% m/v | 5 g |
| EDTA | 0.02 M | 1.46 g |
| Sodium chloride | 1.4 M | 20.45 g |
| TRIS | 0.1 M | 3.028 g |
| Total | n/a | 250 mL |
2. CIA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Chloroform | 96% (v/v) | 48 mL |
| Isoamyl alcohol | 4% (v/v) | 2 mL |
| Total | n/a | 50 mL |
Laboratory supplies
1. 2.0 mL Eppendorf tubes (Citotest, catalog number: 4610-1913)
2. 1.5 mL Eppendorf tubes (Citotest, catalog number:4610-1842)
3. 50 mL Falcon tubes (GenFollower, catalog number: CTB50YE)
4. Pipette tips (Thermo Scientific, model: Finnpipette F1)
Equipment
1. Pipettes (Thermo Scientific, model: Finnpipette F1, catalog numbers: 4641010N, 4641070N, 4641100N, and 4641120)
2. Stainless steel spatula (8") (VWR, catalog number: 82027-532)
3. Mortar (diameter: 100mm) and pestle (size: 117 mm) (DURAN, catalog number: 2120074)
4. High-speed centrifuge (Thermo Scientific, model: MicroCL 21R, catalog number: 75002470)
5. Refrigerator (4°C) (Panasonic Biomedical, model: MPR-721, catalog number: MPR-721-PA)
6. Oven (65°C) (Memmert, model: UN30, catalog number: UN30)
7. Magnetic stirrer (IKA, model: C-MAG HS 7, catalog number: 0020002690)
8. Electrophoresis system (Labnet International, model: ENDUROTM Horizontal Gel Box, catalog number: E1010-E)
9. Photodocumentation system (Labnet International, model: ENDUROTM GDS Touch, catalog number: GDS-1300)
Software and datasets
1. Canvas LMS (Versión 2025.4.10). The chart is available at the following link: https://www.canva.com/design/DAGkQWGQxUM/_eq90lI0a2AZSLoaquijpw/edit?utm_content=DAGkQWGQxUM&utm_campaign=designshare&utm_medium=link2&utm_source=sharebutton
2. RStudio (Posit Software, PBC, © 2009–2024)
3. ImageJ (Fiji, Version 1.52)
Procedure
文章信息
稿件历史记录
提交日期: Jan 22, 2025
接收日期: Mar 30, 2025
在线发布日期: Apr 20, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Riascos-España, A. F., Cuastumal, B. T. A., Zambrano, M. C. I., Arteaga, J. Z. C. and Velasquez-Vasconez, P. A. (2025). Optimized Protocol for DNA Extraction in Three Theobroma Species. Bio-protocol 15(9): e5297. DOI: 10.21769/BioProtoc.5297.
分类
植物科学 > 植物分子生物学 > DNA > DNA 提取
分子生物学 > DNA > DNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














