- EN - English
- CN - 中文
Proteome Birthdating: A Single-Sample Approach for Measuring Global Turnover Dynamics and “Protein Age”
蛋白质组生成年代测定:单样本解析整体蛋白质更新动态与“蛋白质年龄”方法
发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5296 浏览次数: 2203
评审: Emilie BesnardSébastien GillotinAnonymous reviewer(s)

相关实验方案

适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
Basil Baby Mattamana [...] Peter Allen Faull
2025年12月20日 1261 阅读
Abstract
Within a cell, proteins have distinct and highly variable half-lives. As a result, the molecular ages of proteins can range from seconds to years. How the age of a protein influences its environmental interactions is a largely unexplored area of biology. To facilitate such studies, we recently developed a technique termed “proteome birthdating” that differentially labels proteins based on their time of synthesis. Proteome birthdating enables analyses of age distributions of the proteome by tandem mass spectrometry (LC–MS/MS) and provides a methodology for investigating the protein age selectivity of diverse cellular pathways. Proteome birthdating can also provide measurements of protein turnover kinetics from single, sequentially labeled samples. Here, we provide a practical guide for conducting proteome birthdating in in vitro model systems. The outlined workflow covers cell culture, isotopic labeling, protein extraction, enzymatic digestion, peptide cleanup, mass spectrometry, data processing, and theoretical considerations for interpretation of the resulting data.
Key features
• Proteome birthdating barcodes the proteome with isotopically labeled precursors based on time of synthesis or “age.”
• Global protein turnover kinetics can be analyzed from single, sequentially labeled biological samples.
• Protein age distributions of subsets of the proteome can be analyzed (e.g., ubiquitinated proteins).
• Age selectivity of protein properties, cellular pathways, or disease states can be investigated.
Keywords: Protein turnover (蛋白质更新)Graphical overview
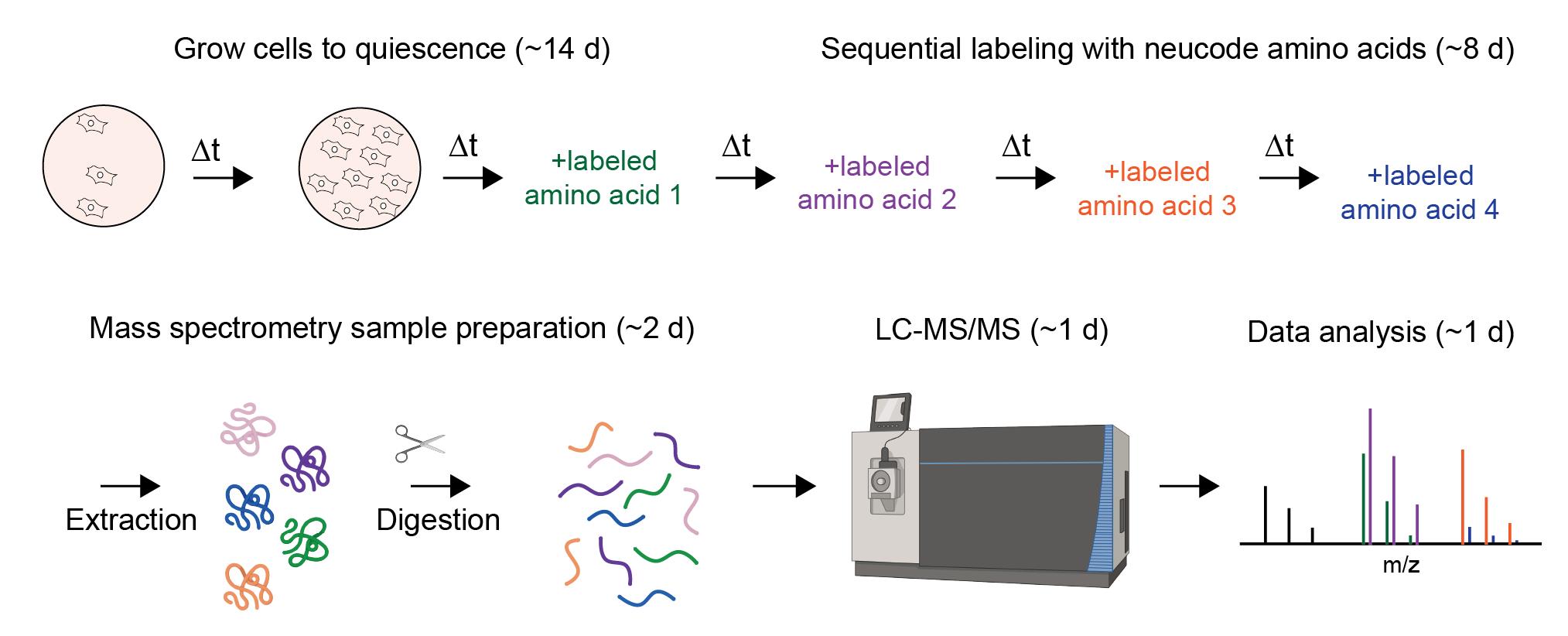
Proteome birthdating: A single-sample approach for measuring global turnover dynamics and “protein age”
Background
Cellular proteins exist in a dynamic equilibrium and are continuously synthesized and degraded. Protein turnover kinetics are highly variable, and protein half-lives can range from minutes to years [1–7]. Accordingly, the "ages" of individual protein molecules within a cell are also highly variable—whereas some cellular proteins have been recently synthesized, others have persisted in the cell for days or even years. As proteins age, their tendency to become misfolded, oxidized, or damaged increases. A protein's age has also been shown to be correlated to post-translational modifications such as ubiquitination [8]. However, investigating how a protein molecule’s age influences its function has been experimentally challenging as most biochemical methodologies cannot differentiate proteins based on their time of synthesis.
A number of recently developed methods have combined time-resolved metabolic labeling with bottom-up proteomics to investigate protein turnover dynamics on global scales [3,9–12]. These approaches, often referred to as “pulsed” or “dynamic” stable isotope labeling by amino acids in cell culture (dSILAC), typically employ a continuous labeling approach wherein an isotopically labeled precursor (an amino acid containing stable heavy isotopes, e.g., 13C615N2 l-lysine) is introduced to cultured cells or whole model organisms over time. Changes in relative levels of newly synthesized proteins harboring the labeled precursor are measured in different experimental time points to determine the kinetics of protein turnover.
Recently, we developed an alternative proteomic approach to dSILAC, named proteome birthdating, that utilizes a sequential rather than continuous labeling strategy [8]. In proteome birthdating, a series of different isotopically labeled precursors are sequentially added to and then removed from the same cell population over time. At the conclusion of the experiment, the relative levels of each label are analyzed at a single experimental endpoint. By taking advantage of neutron-encoded (NeuCode) amino acids [13,14], our initial study was able to "barcode" proteins with five labels, exceeding the multiplexing limit of three that is typical of standard SILAC experiments. NeuCode amino acids are isotopically labeled groups of amino acids with the same number of additional neutrons but different atomic arrangements of those neutrons (e.g., K602 13C615N2 l-lysine vs. K080 2H8 l-lysine; Figure 1). The slight mass differences between NeuCode amino acids allow for multiplexed metabolic labeling in quantitative proteomic experiments while minimizing the loss of proteome coverage [13,14].
The application of proteome birthdating advances proteomic analyses in two ways. First, it provides a methodology for analyzing protein half-lives that offers some advantages over pulsed/dynamic SILAC. Specifically, it allows proteome dynamics to be investigated by analyzing a single biological sample, rather than a series of time points. Second, proteome birthdating provides an approach for partitioning proteins within a cell according to their molecular age, thus facilitating investigations of age-specific protein properties. To demonstrate these two applications of proteome birthdating, we used the approach to measure global protein half-lives in primary human fibroblasts and conducted an age census of the human ubiquitinome [8].
Detailed below is a practical guide for conducting proteome birthdating in cultured cells. The protocol enables measurements of turnover kinetics and “protein age” from single biological samples. Although the protocol specifically applies to human dermal fibroblasts and uses the Thermo Orbitrap Astral LC–MS/MS system, it can be adapted to alternative cultured cell systems and other high-resolution mass spectrometers.
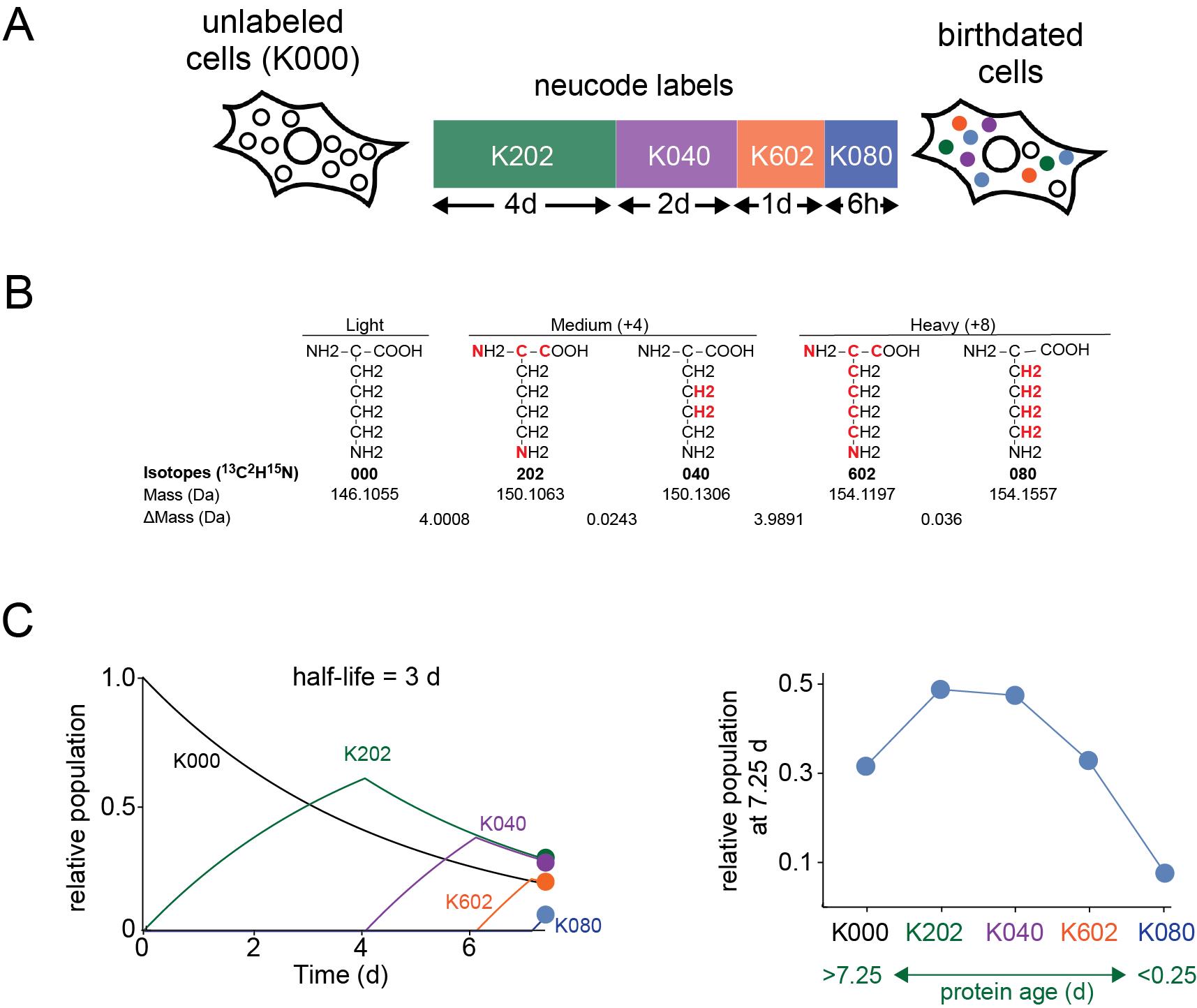
Figure 1. General design of proteome birthdating experiments. (A) In proteome birthdating, multiple isotopically labeled amino acids (in this case, five NeuCode isotopic variants of lysine) are sequentially added and then removed from the same biological sample. Relative levels of differentially labeled variants of proteins are then measured at the endpoint of the experiment. In the KXYZ nomenclature, X, Y, and Z refer to the number of 13C, 2H, and 15N atoms. (B) Structures show the five NeuCode isotopic variants of lysine used in our published proteome birthdating experiments [8]. The red atoms indicate the sites of heavy isotope incorporation. ΔMass refers to the difference in mass between adjacent variants. (C) The kinetic plots show theoretical changes in relative populations of KXYZ-labeled peptides over time during the 7.5-day labeling time course shown in (A) for a theoretical protein having a half-life of 3 days. The right plot shows the relative populations of KXYZ-labeled peptides at the end of the time course. Using this approach, proteins are isotopically labeled based on their time of synthesis (i.e., “age”) with relative populations of labeled forms of a protein being established by its rate of turnover.
Materials and reagents
Biological materials
1. hTERT immortalized HCA2 human fibroblasts or other in vitro model systems
a. Equivalent and well-established hTERT-immortalized human foreskin fibroblasts can be obtained from any commercial repository, e.g., ATCC, #CRL-4001. Detailed instructions on the maintenance and culturing of this cell-type are also available here: https://www.atcc.org/resources/culture-guides/htert-immortalized-cell-culture-guide#Foreskin
b. Primary cell lines with the potential to become quiescent offer the theoretical advantage that labeling can be conducted in a non-dividing state where only protein turnover (and not protein dilution due to cell division) influences labeling kinetics
c. Cell lines lacking the potential to become quiescent can be birthdated. However, if the goal of the experiment is to measure protein turnover kinetics, the rate of cell division of the culture must be accurately measured and subtracted from the observed labeling kinetics (see General notes and troubleshooting section)
Reagents
1. Fetal bovine serum (FBS) (Gibco, catalog number: A5256701)
2. Dialyzed fetal bovine serum (dFBS) (Gibco, catalog number: 26400044)
3. Eagle’s modified essential media (EMEM) (ATCC, catalog number: 30-2003)
4. Minimal essential media for SILAC (MEM for SILAC) (Thermo Scientific, catalog number: 88368)
5. 50 mg/mL Primocin or 10 mg/mL penicillin-streptomycin (Pen-Strep) (Invitrogen, catalog number: 15070063 or Gibco, catalog number: 15140122)
6. Phosphate buffered saline (PBS) (Corning, catalog number: 21-040)
7. Trypsin for tissue culture (Gibco, catalog number: 25200056)
8. L-arginine 000 (R000) (Thermo Fisher, catalog number: 89989 or Cambridge Isotope Laboratories, catalog number: ULM-8347-PK)*
9. L-lysine 000 (K000) (Thermo Fisher, catalog number: 89987 or Cambridge Isotope Laboratories, catalog number: ULM-8766-PK)*
10. NeuCode amino acids
a. L-lysine 202 (K202) (Thermo Fisher, catalog number: A36754)*
b. L-lysine 040 (K040) (Thermo Fisher, catalog number: 88438 or Cambridge Isotope Laboratories, catalog number: DLM-2640-PK)*
c. L-lysine 602 (K602) (Thermo Fisher, catalog number: 88432 or Cambridge Isotope Laboratories, catalog number: CNLM-291-H-PK)*
d. L-lysine 080 (K080) (Thermo Fisher, catalog number: A33613 or Cambridge Isotope Laboratories, catalog number: DLM-2641-PK)*
11. BCA kit (Thermo Scientific, catalog number: 23235)
12. Dithiothreitol (DTT) (Thermo Scientific, catalog number: A39255)**
13. Iodoacetamide (IAA) (Thermo Scientific, catalog number: 35603)**
14. MS water (Thermo Scientific, catalog number: 51140)
15. MS trypsin (Thermo Scientific, catalog number: 90305)*
16. MS Lys-C (Thermo Scientific, catalog number: 90307)*
17. MS 0.1% Trifluoroacetic acid in water (TFA) (Elution buffer 1) (Thermo Scientific, catalog number: LS119)**
18. MS acetonitrile (MeCN) (Thermo Scientific, catalog number: PI51101)**
19. MS 1 M triethylammonium bicarbonate (1 M TEAB) (Thermo Scientific, catalog number: 90114)
20. MS 0.1% formic acid in water (Fisher Scientific, catalog number: T85170K2)**
21. MS 80% acetonitrile, 20% water with 0.1% formic acid (Fisher Scientific, catalog number: LS122500)**
22. 5 N ammonium hydroxide (NH4OH) (Fisher Scientific, catalog number: LC111101)**
23. 100% methanol (Thermo Scientific, catalog number: A412-500)**
24. 20% SDS (Bio-Rad, catalog number: 1610418)
25. 10× radioimmunoprecipitation assay buffer (RIPA) (EMD Millipore, catalog number: 20-188)
26. Pierce protease and phosphatase inhibitor mini tablets (Thermo Scientific, catalog number: A32959)
27. 2× Laemmli sample buffer (Bio-Rad, catalog number: 1610737EDU)
28. 100% phosphoric acid (H3PO4) (Fisher Scientific, catalog number: 02003602)**
*These substances are known to be temperature and moisture sensitive. Please follow all handling guidelines provided by the manufacturer.
**These substances are potentially hazardous. Please ensure to observe the recommended safety and handling guidelines in the Materials and Safety Data Sheet (MSDS) provided by the manufacturer.
Solutions
A. Cell culture media
1. Maintenance media (see Recipes)
2. Adapting media (see Recipes)
3. Labeling media (see Recipes)
B. Sample preparation
4. SDS lysis buffer (see Recipes)
5. RIPA lysis buffer (see Recipes)
6. 25 mM dithiothreitol (DTT) solution (see Recipes)
7. 125 mM iodoacetamide (IAA) solution (see Recipes)
8. S-TRAP wash buffer (see Recipes)
9. 12% phosphoric acid (see Recipes)
10. Digestion buffer (50 mM TEAB) (see Recipes)
11. Elution buffer 2 (0.05% TFA in 50% MeCN) (see Recipes)
12. Fractionation buffers (see Recipes)
C. LC–MS/MS solutions
14. HPLC solvent A (0.1% FA in water) (see Reagents)
15. HPLC solvent B (0.1% FA in 80% ACN) (see Reagents)
Recipes
1. Maintenance media (500 mL)
Note: Media can be sterile-filtered after mixing and stored at 4 °C for 2 weeks.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EMEM | 85% v/v | 425 mL |
| FBS | 15% v/v | 75 mL |
| 50 mg/mL Primocin | 100 μg/mL | 1 mL |
| Alternative: 10 mg/mL Pen-Strep | 100 μg/mL | 5 mL |
2. Adapting media (500 mL)
Note: Media can be sterile-filtered after mixing and stored at 4 °C for 2 weeks.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EMEM | 85% v/v | 425 mL |
| Dialyzed FBS | 15% v/v | 75 mL |
| 50 mg/mL Primocin | 100 μg/mL | 1 mL |
| Alternative: 10 mg/mL Pen-Strep | 100 μg/mL | 5 mL |
3. Labeling media (500 mL)
Note: Media can be sterile-filtered after mixing and stored at 4 °C for 2 weeks.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MEM for SILAC | 85% v/v | 425 mL |
| 50 mg/mL Primocin | 100 μg/mL | 1 mL |
| Alternative: 10 mg/mL Pen-Strep | 100 μg/mL | 5 mL |
| Dialyzed FBS | 15% v/v | 75 mL |
| Arginine 000 | 750 μM | 65 mg |
| Variant 1: Lysine 000 | 620 μM | 45.2 mg |
| Variant 2: Lysine 202 | 620 μM | 45.2 mg |
| Variant 3: Lysine 040 | 620 μM | 45.2 mg |
| Variant 4: Lysine 602 | 620 μM | 45.2 mg |
| Variant 5: Lysine 080 | 620 μM | 45.2 mg |
4. SDS lysis buffer
Note: This recipe is meant for harsh lysis; for gentler lysis, please see Recipe 5. Prepare at room temperature. Can be stored at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20% SDS | 5% v/v | 250 μL |
| 1 M TEAB | 50 mM | 50 μL |
| MS Water | 70% v/v | 700 μL |
5. RIPA lysis buffer
Note: This recipe is meant for gentle lysis; for harsher lysis, please see Recipe 4. Prepare on ice. Can be stored at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10× RIPA buffer | 10% v/v | 1 mL |
| Protease/phosphatase inhibitor tablet | 1× | 1 tablet |
| MS water | 90% v/v | 9 mL |
6. 25 mM dithiothreitol (DTT) solution
Note: Prepare fresh. This is a potentially hazardous recipe. Ensure safety measures are followed.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DTT | 25 mM | 3.85 mg |
| 1 M TEAB | 50 mM | 50 μL |
| MS water | 95% v/v | 950 μL |
7. 125 mM iodoacetamide (IAA) solution
Note: Prepare fresh and protect from light. This is a potentially hazardous recipe. Ensure safety measures are followed.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| IAA | 125 mM | 21.8 mg |
| 1 M TEAB | 50 mM | 50 μL |
| MS water | 95% v/v | 950 μL |
8. S-TRAP wash buffer
Note: Prepare final solution fresh and store the stock solution of 1 M TEAB at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M TEAB | 100 mM | 1 mL |
| 100% methanol | 90% v/v | 9 mL |
9. 12% phosphoric acid
Note: Can be prepared and stored at room temperature. This is a potentially hazardous recipe. Ensure safety measures are followed.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 100% H3PO4 | 12% v/v | 120 μL |
| MS water | 88% v/v | 880 μL |
10. Digestion buffer (50 mM TEAB)
Note: Prepare final solution fresh and store the stock solution of 1 M TEAB at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M TEAB | 50 mM | 100 μL |
| MS water | 95% v/v | 1,900 μL |
| MS trypsin | 0.05 μg/μL | 100 μg |
| Alternative: MS Lys-C | 0.05 μg/μL | 100 μg |
11. Elution buffer 2 (0.05% TFA in 50% MeCN)
Note: This is a potentially hazardous recipe. Ensure safety measures are followed.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 0.1% MS TFA in water | 0.05% v/v | 1 mL |
| MS MeCN | 50% v/v | 1 mL |
12. Fractionation buffers
Note: This is a potentially hazardous recipe. Ensure safety measures are followed.
| Fraction # | Final % MeCN | 100% MeCN μL | 10 mM NH4OH μL |
| Fractionation wash buffer | 0 | 0 | 1,000 |
| E1 | 2 | 20 | 980 |
| E2 | 3.5 | 35 | 965 |
| E3 | 5 | 50 | 950 |
| E4 | 6.5 | 65 | 935 |
| E5 | 8 | 80 | 920 |
| E6 | 9.5 | 95 | 905 |
| E7 | 11 | 110 | 890 |
| E8 | 12.5 | 125 | 875 |
| E9 | 14 | 140 | 860 |
| E10 | 15.5 | 155 | 845 |
| E11 | 17 | 170 | 830 |
| E12 | 18.5 | 185 | 815 |
| E13 | 20 | 200 | 800 |
| E14 | 21.5 | 215 | 785 |
| E15 | 27 | 270 | 730 |
| E16 | 50 | 500 | 500 |
Laboratory supplies
1. MS-compatible pipettes (any brand)
2. MS-compatible pipette tips (any brand)
3. Low-bind 2 mL tubes (Eppendorf, catalog number: 022431102)
4. Flat-bottom, clear 96-well plate (Grenier, catalog number: 655101)
5. Tissue-culture serological pipettes (any brand)
6. S-TRAP micro columns (Protifi, catalog number: C02-micro-10)
7. C18 StageTips for desalting and fractionation (Fisher Scientific, catalog number: 13110055)
8. SureSTART autosampler vials (Thermo Fisher, catalog number: 6ERV11-03PPC)
9. SureSTART autosampler vial caps (Thermo Fisher, catalog number: 6ARC11NRT1)
10. Acclaim PepMap 100 C18 HPLC trap column, 20 mm × 2 cm, 3 μm (Thermo Fisher, catalog number: 164946)
11. nanoViper fitting transfer column for Vanquish Neo, double nanoViper, 0.020 × 550 mm (1,500 bar) (Thermo Fisher, catalog number: 6250.5260)
12. nanoViper fitting column for Vanquish Neo, double nanoViper, 0.050 × 150 mm (1,500 bar) (Thermo Fisher, catalog number: 6250.5124)
13. Needle seat for Vanquish (Thermo Fisher, catalog number: 6252.2470)
14. Sample loop for Vanquish split sample NT, biocompatible, 25 μL (Thermo Fisher, catalog number: 6252.1940)
15. Aurora Rapid 8 × 75 XT C18 UHPLC column (IonOpticks, catalog number: AUR3-8075C18-XT)
Equipment
1. CentriVap Vacuum Concentrator (Labconco)
2. Cell culture hood (Labconco)
3. Cell culture incubator (Thermo Fisher)
4. Waste vacuum (any brand)
5. Benchtop centrifuge capable of 19,000× g and 4 °C (any brand)
6. Benchtop sample rotator (any brand)
7. Benchtop vortexer (any brand)
8. Heat block (any brand)
9. Plate reader capable of reading absorbance at 562 nm (SpectraMax)
10. Non-tissue culture incubator (any brand)
11. IonOpticks heater controller (IonOpticks, catalog number: IOHEATCON1)
12. IonOpticks column heater (IonOpticks, catalog number: COLHTR01)
13. Vanquish Neo UHPLC system (Thermo Fisher, catalog number: VN-S10-A-01)
14. Orbitrap Astral mass spectrometer (Thermo Fisher, catalog number: BRE725600)
15. EASY-spray source (Thermo Fisher, catalog number: ES081)
Software and datasets
1. MSFragger v4.0 (FragPipe v21.1 with Philosopher v5.1.1)
2. UniProtKB (https://www.uniprot.org/help/uniprotkb)
3. Orbitrap Astral Tune Application Version 1.1.477.46 (Thermo Fisher)
4. Chromeleon Console Version 7.3.2.10767 (Thermo Fisher)
5. XCalibur Version 4.7.0.105 (Thermo Fisher)
6. ProteoWizard MS Convert (https://proteowizard.sourceforge.io/)
7. Wolfram Mathematica v14.1(Mathematica notebooks used for custom analysis, data modeling, and graphing have been archived on Zenodo at https://doi.org/10.5281/zenodo.10866478
Procedure
文章信息
稿件历史记录
提交日期: Jan 17, 2025
接收日期: Apr 1, 2025
在线发布日期: Apr 20, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Meadow, M. E., Broas, S., Hoare, M., Ahmed, M., Alimohammadi, F., Welle, K. A., Swovick, K., Hryhorenko, J. R., Jain, A., Martinez, J. C., Seluanov, A., Gorbunova, V., Buchwalter, A. and Ghaemmaghami, S. (2025). Proteome Birthdating: A Single-Sample Approach for Measuring Global Turnover Dynamics and “Protein Age”. Bio-protocol 15(9): e5296. DOI: 10.21769/BioProtoc.5296.
分类
系统生物学 > 蛋白质组学
细胞生物学 > 基于细胞的分析方法 > 蛋白成熟
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











