- EN - English
- CN - 中文
snPATHO-seq: A Detailed Protocol for Single Nucleus RNA Sequencing From FFPE
snPATHO-seq:基于FFPE样本的单核转录组测序详细方法
(*Contributed equally to this work, §Contributed equally to this work) 发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5291 浏览次数: 1961
评审: Alessandro DidonnaAnonymous reviewer(s)
Abstract
Formalin-fixed paraffin-embedded (FFPE) samples remain an underutilized resource in single-cell omics due to RNA degradation from formalin fixation. Here, we present snPATHO-seq, a robust and adaptable approach that enables the generation of high-quality single-nucleus (sn) transcriptomic data from FFPE tissues, utilizing advancements in single-cell genomic techniques. The snPATHO-seq workflow integrates optimized nuclei isolation with the 10× Genomics Flex assay, targeting short RNA fragments to mitigate FFPE-related RNA degradation. Benchmarking against standard 10× 3' and Flex assays for fresh/frozen tissues confirmed robust detection of transcriptomic signatures and cell types. snPATHO-seq demonstrated high performance across diverse FFPE samples, including diseased tissues like breast cancer. It seamlessly integrates with FFPE spatial transcriptomics (e.g., FFPE Visium) for multi-modal spatial and single-nucleus profiling. Compared to workflows like 10× Genomics’ snFFPE, snPATHO-seq delivers superior data quality by reducing tissue debris and preserving RNA integrity via nuclei isolation. This cost-effective workflow enables high-resolution transcriptomics of archival FFPE samples, advancing single-cell omics in translational and clinical research.
Key features
• Optimized nuclei isolation from FFPE tissues enables high-quality single-nucleus transcriptomics by minimizing debris and maximizing intact nuclear yield.
• Compatible with 10× Genomics Flex, leveraging short RNA probes to overcome FFPE RNA fragmentation challenges.
• Outperforms existing FFPE workflows in cell type detection sensitivity across archival, degraded, or aged samples.
• Low-cost, accessible protocol using off-the-shelf reagents, suitable for broad translational and archival tissue applications.
Keywords: snRNA-seq (单核RNA测序)Graphical overview
Background
Clinical tissue specimens are predominantly preserved using the formalin-fixed paraffin-embedded (FFPE) technique, a universally established method for the long-term conservation of biological tissues. In the United States alone, pathology laboratories process between 10,000 and 100,000 FFPE blocks annually, underscoring the immense scale and significance of this resource for clinical diagnostics and translational research [1]. These FFPE samples serve as a cornerstone for retrospective studies, enabling molecular and histopathological analyses that drive advancements in precision medicine and biomedical discovery [1]. Advances in molecular technologies, including genomics, transcriptomics, and proteomics, have expanded the utility of FFPE samples for molecular characterization [2,3]. However, these applications are traditionally conducted at the bulk tissue level, which fails to resolve the intricate molecular heterogeneity inherent in cancers and other complex biological settings. Technologies enabling single-cell transcriptomic profiling of FFPE tissues thus hold immense potential for advancing human health research.
Previously, our team successfully conducted single-nucleus (sn) transcriptomic profiling using clinical human FFPE tissue samples, demonstrating the feasibility of isolating intact nuclei from FFPE samples for molecular characterization [4]. However, extending this technology to transcriptomics poses a significant challenge due to RNA fragmentation caused by formalin fixation, heat exposure, and paraffin embedding [2]. RNA degradation poses a significant challenge to the performance of widely used single-cell/nucleus RNA sequencing (sc/sn RNA-seq) methods, such as the 10× Genomics 3' assay and SMART-seq, which rely on poly(dT) probes to selectively capture and reverse-transcribe intact mRNA molecules [5,6]. RNA-binding probes enable gene expression profiling in FFPE samples and are used in spatial methods like MERFISH, 10× Visium, and Xenium. While resilient to RNA fragmentation, these methods lack transcriptome-wide or single-cell resolution [7–9]. To address this limitation, 10× Genomics introduced Flex chemistry, which employs RNA-targeting probes capable of capturing short RNA fragments, significantly improving compatibility with FFPE-derived material [10].
To address challenges in analyzing FFPE samples, we developed snPATHO-seq, a nuclei isolation protocol optimized for FFPE tissues. This workflow integrates rehydration, enzyme-based dissociation, and nuclei isolation (an example of good-quality nuclei is shown in Figure 1), followed by gene expression analysis using the 10× Genomics Flex assay. We validated snPATHO-seq on three human breast cancer FFPE samples, demonstrating strong agreement with standard 10× 3' and Flex chemistries, previously restricted to fresh or frozen tissues.
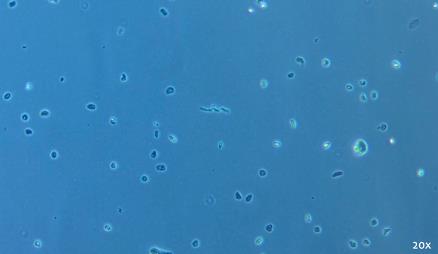
Figure 1. Representative example of good-quality nuclei preparation. Brightfield image was taken with a 20× objective on the ZEISS Primovert.
In parallel, 10× Genomics introduced a single-cell fixation protocol for single-cell RNA sequencing [11]. 10× Genomics’ scFFPE protocol does not include a defined nuclei isolation step, leading to the isolation of a mixture of intact cells and nuclei from FFPE samples. This incomplete separation can compromise data quality by introducing variability in transcript capture efficiency and downstream interpretation. In contrast, snPATHO-seq incorporates dedicated membrane lysis and nuclei isolation steps, enabling consistent enrichment for nuclei and minimizing contamination from partially digested cells. Our direct comparison of the two workflows demonstrated that snPATHO-seq delivers more robust and reproducible gene expression profiles, better suited for high-quality single-nucleus transcriptomics from FFPE tissues [12]. This technical advancement strengthens snPATHO-seq as a more mature and reliable protocol for FFPE single-nucleus studies.
In summary, snPATHO-seq unlocks the potential of archival FFPE samples, offering a robust and mature protocol for single-nucleus transcriptomic analysis. By overcoming the technical challenges of RNA degradation and enabling single-cell resolution profiling, snPATHO-seq bridges the gap between traditional pathology and molecular research, fostering new opportunities for clinical and translational studies.
Materials and reagents
Biological materials
1. FFPE tissue archive blocks
Reagents
1. RNAse Away surface decontaminant (Thermo Fisher Scientific, catalog number: 7002PK)
2. Xylene (Sigma, catalog number: 214736)
3. Ethanol (Sigma, catalog number: E7023)
4. RPMI-1640 media (Gibco, catalog number: 11875093)
5. Liberase TH 100 mg (Sigma, catalog number: 5401135001)
6. RiboLock RNase inhibitor (40 U/μL) (Thermo Fisher Scientific, catalog number: EO0382)
7. Nuclei Isolation kit Nuclei EZ Prep (Sigma, catalog number: NUC101)
8. Bovine serum albumin (BSA) solution, sterile filtered (Sigma-Aldrich, catalog number: A1595)
9. LiberaseTM research grade, 10 mg (Roche, catalog number: 5401119001)
10. Collagenase D (Roche, catalog number: 11088858001)
11. 10× PBS buffer, pH 7.4 (Invitrogen, catalog number: AM9624 or AM9625)
12. Ambion nuclease-free water (Invitrogen, catalog number: AM9932)
13. Acridine Orange/Propidium Iodide (AO/PI) Viability kit (Logos, catalog number: F23011)
Solutions
1. 70% Ethanol (see Recipes)
2. 50% Ethanol (see Recipes)
3. 30% Ethanol (see Recipes)
4. Digestion solution (see Recipes)
5. Lysis solution (see Recipes)
6. Wash solution 1 (see Recipes)
7. Wash solution 2 (see Recipes)
Recipes
1. 70% Ethanol
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Ethanol | 100% | 70% | 770 | 1540 |
| Nuclease-free water | - | - | 330 | 660 |
| Total | 1,100 | 2,200 |
2. 50% Ethanol
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Ethanol | 100% | 50% | 550 | 1,100 |
| Nuclease-free water | - | - | 550 | 1,100 |
| Total | 1,100 | 2,200 |
3. 30% Ethanol
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Ethanol | 100% | 30% | 330 | 660 |
| Nuclease-free water | - | - | 770 | 1,540 |
| Total | 1,100 | 2,200 |
4. Digestion solution
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Liberase TH | 1 mg/mL | 1 mg/mL | 1072.5 | 2,145 |
| RNAse inhibitor | 40 U/μL | 1 U/μL | 27.5 | 55 |
| Total | 1,100 | 2,200 |
Note: 1 mg/mL of Liberase TH works well for most tissues we tested, but testing different concentrations up to 5 mg/mL for your tissue type is recommended.
5. Lysis solution
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Nuclei EZ lysis buffer | 1× | 1× | 825 | 1,650 |
| BSA | 10% | 2% | 220 | 440 |
| RNAse inhibitor | 40 U/μL | 1 U/μL | 55 | 110 |
| Total | 1,100 | 2,200 |
6. Wash solution 1
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Nuclease-free water | - | - | 1,760 | 3,520 |
| PBS | 10× | 1× | 220 | 440 |
| BSA | 10% | 1% | 220 | 440 |
| Total | 2,200 | 4,400 |
7. Wash solution 2
| Reagent | Stock | Final | 1× + 10% (μL) | 2× + 10% (μL) |
|---|---|---|---|---|
| Nuclease-free water | - | - | 2,085.6 | 4,171.2 |
| PBS | 10× | 0.5× | 110 | 220 |
| BSA | 10% | 0.02% | 4.4 | 8.8 |
| Total | 2,200 | 4,400 |
Laboratory supplies
1. Desiccant silica gel packets (Root Lab, catalog number: DES100)
2. Bel-Art disposable pestles (Sigma, catalog number: BAF199230001)
3. Precision glide needle 25 G (BD, catalog number: 962776)
4. 1 mL syringe (Sigma, catalog number: Z683531-100EA)
5. DNA LoBind tubes 1.5 mL (Eppendorf, catalog number: 022431021)
6. DNA LoBind tubes 2.0 mL (Eppendorf, catalog number: 022431048)
7. pluriStrainer mini 70 μm (pluriSelect, catalog number: 43-10070-40)
8. pluriStrainer mini 40 μm (pluriSelect, catalog number: 43-50040-51)
9. Falcon conical centrifuge tubes (Corning, catalog numbers: 352070 for 50 mL, 352095 for 15 mL)
10. Sterile serological pipettes with pipettor
11. Luna Photon low fluorescent counting chambers (Logos Biosystems, catalog number: L12005)
12. Rainin Pipet-Lite LTS filtered pipette tips (Rainin, catalog numbers: 30389240, 30389213, 30389226)
Equipment
1. HistoCore BIOCUT, manual rotary microtome (Leica, catalog number: 149BIO000C1)
2. Digital dry bath/block heater (Thermo Fisher, catalog number: 88870001)
3. ThermoMixer C (Eppendorf, catalog number: 5382000023)
4. Centrifuge 5810/5810R (Eppendorf, catalog number: EP022628188) with rotor S-4-104
5. Rotor F-35-6-30 (Eppendorf, catalog number: EP5427716009 or similar)
6. S-4-104 rotor adapters for 50× 1.5/2 mL tubes (Eppendorf, catalog number: 58-257-40009)
7. Standard heavy-duty vortex mixer (VWR or Fisherbrand, catalog number: 97043-562)
8. Ice bucket with cool blocks or cool rack CFT30 (Corning, catalog number: CLS432052)
9. Luna-FX7 automated cell counter (Logos Biosystems, catalog number: L700002)
Software and datasets
1. BioRender (https://www.biorender.com/). The following figures were created using BioRender: Graphical overview, https://BioRender.com/j4ty88o
Procedure
文章信息
稿件历史记录
提交日期: Feb 3, 2025
接收日期: Mar 31, 2025
在线发布日期: Apr 11, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Arjumand, W., Wise, K., DuBose, H., Plummer, J. and Martelotto, L. G. (2025). snPATHO-seq: A Detailed Protocol for Single Nucleus RNA Sequencing From FFPE. Bio-protocol 15(9): e5291. DOI: 10.21769/BioProtoc.5291.
分类
细胞生物学 > 单细胞分析
系统生物学 > 转录组学 > RNA测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link














