- EN - English
- CN - 中文
Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
基于Fiji ImageJ的全自动化流程开发:批量分析共聚焦图像数据并量化蛋白共定位的Manders系数
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5285 浏览次数: 2879
评审: Pilar Villacampa AlcubierreXiaochen SunRobert KiewiszKerui Huang
Abstract
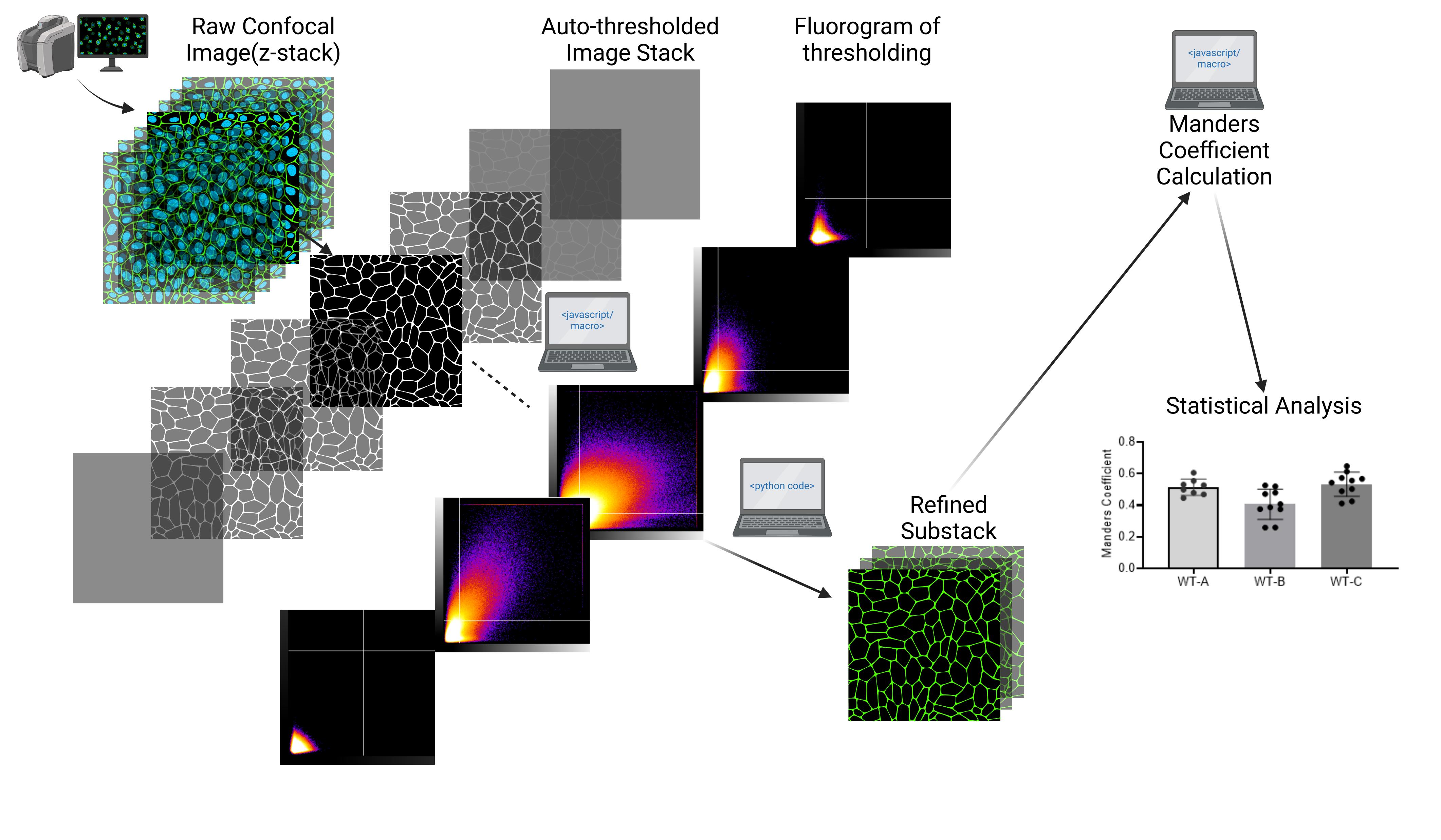
Confocal microscopy is integral to molecular and cellular biology, enabling high-resolution imaging and colocalization studies to elucidate biomolecular interactions in cells. Despite its utility, challenges in handling large datasets, particularly in preprocessing Z-stacks and calculating colocalization metrics like the Manders coefficient, limit efficiency and reproducibility. Manually processing large numbers of imaging data for colocalization analysis is prone to observer bias and inefficiencies. This study presents an automated workflow integrating Python-based preprocessing with Fiji ImageJ's BIOP-JACoP plugin to streamline Z-stack refinement and colocalization analysis. We generated an executable Windows application and made it publicly available on GitHub (https://github.com/weiyue99/Yue-Colocalization), allowing even those without Python experience to directly run the Python code required in the current protocol. The workflow systematically removes signal-free Z-slices that sometimes exist at the beginning and/or end of the Z-stacks using auto-thresholding, creates refined substacks, and performs batch analysis to calculate the Manders coefficient. It is designed for high-throughput applications, significantly reducing human error and hands-on time. By ensuring reproducibility and adaptability, this protocol addresses critical gaps in confocal image analysis workflows, facilitating efficient handling of large datasets and offering broad applicability in protein colocalization studies.
Key features
• Automated image analysis: use of Python-based code for substack creation based on auto-thresholding to eliminate observer bias.
• Manders coefficient calculation: quantification of colocalization using BIOP-JACoP in Fiji ImageJ.
• High-throughput compatibility: efficient for large datasets using macro codes to run in batch mode; minimal manual intervention.
Keywords: Confocal microscopy (共聚焦显微镜)Graphical overview
Created in BioRender. Aditya, V. (2025) https://BioRender.com/m89i580
Background
Confocal microscopy is a cornerstone of modern molecular and cell biology research, providing high-resolution imaging capabilities and generating large volumes of data, especially when performing Z-stack imaging. For example, systems like the Olympus FV10i-LIV or Zeiss LSM series can automatically produce hundreds of images per sample, enabling detailed spatial and temporal studies of biological specimens [1,2]. Among the many applications of confocal microscopy, colocalization analysis is a widely used technique to assess the spatial association between biomolecules, helping to infer protein interactions, co-occurrence, and functional relationships [3]. The Manders coefficient is a commonly employed metric for colocalization studies, offering a quantitative measure of overlap between two signals in fluorescence microscopy datasets [4]. Fiji ImageJ’s Bioimaging and Optics Platform (BIOP) Just Another Colocalization Plugin (JACoP) has been developed for colocalization analysis on confocal images by using the Manders coefficient method [5,6].
Despite its importance, conducting accurate and efficient colocalization analysis remains challenging due to the lack of automated workflows to handle the substantial datasets generated by confocal microscopy. To the best of our knowledge, no comprehensive, automated solution exists in the literature to address these issues, creating an urgent and unmet need for a step-by-step protocol for automated workflows. The primary challenge lies in the preprocessing of Z-stack images, which often include blank slices at the beginning and end of the stack due to the nature of sample preparation and imaging conditions. Including these signal-free slices in Manders coefficient calculations may disrupt thresholding accuracy and lead to inconsistent results [3]. Manually identifying and excluding these blank images is not only time-consuming but also prone to observer bias, which can introduce errors and inconsistencies across datasets.
The current protocol addresses these limitations by providing a detailed, step-by-step automated workflow that integrates a Python-based script to refine raw confocal images. The script identifies and removes signal-free Z-slices, creating refined substacks for further analysis. By coupling this preprocessing step with Fiji ImageJ’s BIOP-JACoP plugin for Manders coefficient calculations, the protocol ensures reliable and reproducible colocalization measurements. This automated approach minimizes human intervention, enhances the accuracy of results, and significantly improves the efficiency of high-throughput studies.
Organic anion transport polypeptide (OATP) 1B1 is a plasma membrane transport protein that plays an important role in drug disposition [7]. Plasma membrane localization of OATP1B1 has often been assessed by quantification of its colocalization with sodium-potassium ATPase (Na/K-ATPase), a plasma membrane protein marker, as we previously published [8,9]. The current protocol uses colocalization of transiently transfected FLAG-tagged OATP1B1 with endogenous Na/K-ATPase in human embryonic kidney (HEK) 293 cells from confocal images as an example, as shown in our original publication [9].
Materials and reagents
Biological materials
1. HEK293 cells (American Type Culture Collection, CRL-1573)
2. pcDNA3.1(+)-WT-OATP1B1: pcDNA3.1 (+) vector-based mammalian expression plasmid vector with the open reading frame of wild-type (WT) OATP1B1 was subcloned at GenScript Biotech (Piscataway, NJ) and verified by sequencing, as described in our publication [9]
Reagents
1. Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, catalog number: 11668019)
2. Antibodies
Primary antibodies: Rabbit FLAG polyclonal antibody (Sigma-Aldrich, catalog number: F7425) and mouse monoclonal Na/K-ATPase antibody (Santa Cruz, catalog number: sc-21712)
Secondary antibodies: Alexa Fluor 594 goat anti-rabbit IgG (Thermo Fisher Scientific, catalog number: A-11012) and Alexa Fluor® 488 goat anti-mouse IgG antibody (Thermo Fisher Scientific, catalog number: A-11001)
3. 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Thermo Fisher Scientific, catalog number: D1306)
Equipment
1. Fluoview microscope (Olympus, model: FV10i-LIV Confocal Imaging System with Live-Cell)
2. Computer system (Windows 10 Enterprise, Processor: 12th Gen Intel® CoreTM i5-12500, 3.00 GHz, RAM: 16.0 GB)Note: Any computer system capable of running Fiji ImageJ on supported platforms is acceptable.
Software and datasets
1. Fiji ImageJ (1.54f, 09/15/2024) [6], open source (https://imagej.net/Fiji/Downloads)
2. BIOP-JACoP (v2.1.1, 08/20/2010) installed to Fiji ImageJ:
Download JACoP from https://github.com/fabricecordelieres/IJ-Plugin_JACoP/releases/download/v2.1.4/JACoP_.jar. Launch Fiji, Help → Update → manage update sites. In the search box, type in PTBIOP → Apply and close → Apply changes → Close.
3. Python (v3.12.6, 09/06/2024), open source (https://www.python.org/downloads/)
4. GraphPad Prism (10.4.1, 12/05/2024), requires subscription (RRID:SCR_002798, https://www.graphpad.com)
4. All code and executable Windows applications have been deposited to GitHub and are publicly available: https://github.com/weiyue99/Yue-Colocalization (10/4/2024).
Procedure
文章信息
稿件历史记录
提交日期: Jan 13, 2025
接收日期: Mar 18, 2025
在线发布日期: Apr 1, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Aditya, V., Tambe, V. and Yue, W. (2025). Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient. Bio-protocol 15(7): e5285. DOI: 10.21769/BioProtoc.5285.
分类
生物信息学与计算生物学
细胞生物学 > 细胞成像 > 共聚焦显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













