- EN - English
- CN - 中文
A Miniaturized Percoll Gradient Method for Isolation of Quiescent Cells of Yeast
基于微型Percoll密度梯度的酵母静息细胞分离方法
(*contributed equally to this work) 发布: 2025年04月20日第15卷第8期 DOI: 10.21769/BioProtoc.5279 浏览次数: 1489
评审: Fernando A Gonzales-ZubiateLucy XieIsmail Tahmaz
Abstract
Quiescence, the temporary and reversible exit from proliferative growth, is a fundamental biological process. Budding yeast is a preeminent model for studying cellular quiescence owing to its rich experimental toolboxes and evolutionary conservation across eukaryotic pathways and processes that control quiescence. Yeast quiescent cells are reported to be isolated by the continuous linear Percoll gradient method and identified by combining different features such as cell cycle, heat resistance, and cell morphology (single cell). Generally, 10–25 mL of Percoll isotonic solution is first obtained by mixing Percoll with NaCl in 12.5–30 mL centrifugal tubes. Then, the gradient is prepared at high speed for 15–60 min. Finally, approximately 2 × 109 cells are collected, overlaid onto the preformed gradient, and centrifuged to obtain distinct cell fractions. This method requires more reagents and samples and special centrifuges and centrifuge tubes. Besides the cost, it is less favorable for experiments that require high-throughput analyses with a small volume of sample each time. The protocol described here aims to solve those problems by combining the use of 2 mL centrifugal tubes with density marker beads. The protocol also focuses on how to optimize the buoyant density distribution of the density gradient solution such that the density bands better match those of different fraction cells. This will help fully separate quiescent and non-quiescent cells. The protocol can be easily adapted to a wide variety of unicellular microbes with different buoyancy density differentiation during cultivation, such as yeast and bacteria.
Key features
• Minimized Percoll gradient solution volume by using a 2 mL centrifugal tube.
• Reduced material and sample consumption and requirements for centrifuges and tubes.
• Optimized buoyant density distribution of density gradient solution with the emphasis on Percoll concentration and centrifugation parameters by using density marker beads as indicators.
• Easily adaptable to a wide variety of unicellular microbes with different buoyancy density differentiation during cultivation.
Keywords: Unicellular microbe (单细胞微生物)Graphical overview
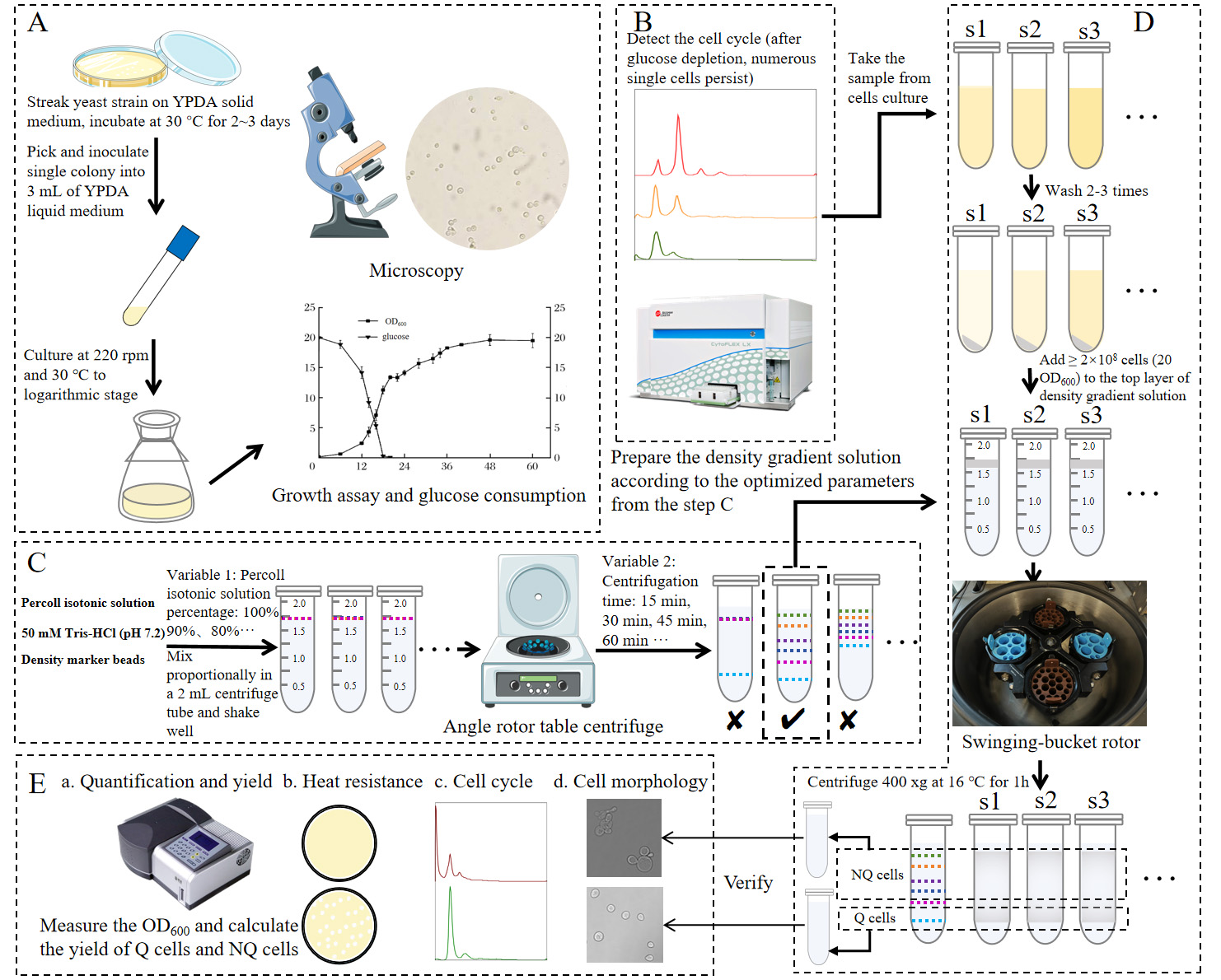
Graphical overview of isolating quiescent cells of yeast using a miniature Percoll gradient method. (A) Strain activation, inoculation, regular sampling, and detection. (B) Cell cycle detection by flow cytometer to determine the time to separate the Q cells. (C) Optimization of Percoll gradient solution preparation to obtain suitable buoyant density distribution in a 2 mL centrifuge tube by using density marker beads. (D) Preparation of density gradient solution and cell suspensions and centrifugation and isolation of Q and NQ cells. (E) Analysis of pre-separation and post-separation cells.
Background
As the temporary and reversible arrest of cell growth, quiescence is a fundamental biological process [1]. The phenomenon of quiescence is commonly present in unicellular microbes, being crucial for their long-term survival under unfavorable conditions [1]. Among them, the budding yeast is a preeminent model system for studying cellular quiescence owing to its rich experimental toolboxes and evolutionary conservation across eukaryotes of pathways and processes that control quiescence [2]. Yeast cell buoyant density has been reported since as early as 1984 [3,4], but its role was determined in 2006 as one of the most important phenotypes of yeast quiescent cells [5]. Heterogeneity within stationary-phase yeast populations was first described by Allen et al. [5]. Two subpopulations were isolated by density centrifugation, and differentiation into quiescent (Q) and non-quiescent (NQ) cells was proposed [5]. Subsequently, the density gradient centrifugation method was followed and used by many other researchers, and more phenotypes of quiescence were reported [6–10]. Generally, 10–25 mL of 90% Percoll solution is obtained by diluting Percoll with 1.5 M NaCl in a 12.5–30 mL centrifugal tube; then, a continuous linear gradient is prepared at high speed for 15–60 min. Finally, approximately 2 × 109 cells (200 OD600) are pelleted, resuspended, overlaid onto the preformed gradient, and centrifuged at 300–400× g for 60 min in a tabletop centrifuge equipped with a swinging bucket rotor. The quiescence phenotypes and corresponding detection methods reported include cell buoyancy density, cell cycle, thermotolerance, cell morphological changes, cell wall thickening, Q phase–related gene detection, cell survival, and synchronous mitotic re-entry capability [5,6,8–10]. In fact, quiescence phenomena are so complex that there is still considerable inconsistency in the methods to identify Q cells among different research groups [1]. Inevitably, it needs multi-faceted or multi-parameter evaluation. Unfortunately, information in those experimental details is not yet adequate, and reporting also lacks standardization [1].
Considering that quiescence phenomena are complex and studies often require multi-sample, multi-timepoint sampling designs, the present isolating method is established to save time and reduce consumable costs by combining a 2 mL centrifugal tube with density marker beads. The key is to optimize the buoyant density distribution of the density gradient solution by investigating the effect of several factors, while also ensuring that the density bands that match Q and NQ cells are formed and fully separated from each other. Further, the cell cycle is selected as the most important and main criterion to determine when quiescent cells emerge and can thus be isolated. With the details fully described here, this protocol would be easily adapted to yeast and other unicellular microbes with similar buoyant cell density range [11,12].
Materials and reagents
Biological materials
1. Yeast strain W303-1A (MATa, ade2, trp1, his3, can1, ura3, leu2), ATCC 208352
Reagents
1. Yeast extract (Sangon Biotech, catalog number: A610961-0500)
2. Peptone A (meat peptone, from bovine) (Sangon Biotech, catalog number: A610213-0500)
3. Agar A (Sangon Biotech, catalog number: A600010-0500)
4. D-(+)-Glucose (Sangon Biotech, catalog number: A501991-0500)
5. Adenine (Sangon Biotech, catalog number: A600013-0100)
6. Ethanol, 99.9% (Tianjin Jiangtian Chemical Technology, catalog number: 19020)
7. Citric acid trisodium dihydrate (C6H5Na3O7·2H2O) (AMRESCO, catalog number: 0101-1KG)
8. Citric acid (C6H8O7) (Sangon Biotech, catalog number: CB0055)
9. SYTOXTM green nucleic acid stain (Thermo Fisher Scientific, catalog number: S7020)
10. RNase A from bovine pancreas (Sangon Biotech, catalog number: B600473-0025)
11. Proteinase K (Transgen, catalog number: GE201-01)
12. PercollTM solution (GE Healthcare, catalog number: 17-0891-09)
13. Sodium chloride (NaCl) (AMRESCO, catalog number: X190 7647-14-5)
14. Tris (Sangon Biotech, catalog number: A600194-0500)
15. Density marker bead (DMB-kit) (Cospheric, catalog number: 5613)
16. DMSO (CALEDON, catalog number: 4100-1-05)
17. Hydrochloric acid (DUKSAN, catalog number: 2065)
Solutions
1. 70% ethanol (see Recipes)
2. Normal saline (see Recipes)
3. 1.5 M NaCl solution (see Recipes)
4. SYTOXTM green working solution (see Recipes)
5. Percoll isotonic solution (see Recipes)
6. YPDA culture medium (see Recipes)
7. Density marker beads (see Recipes)
8. 50 mM sodium citrate buffer (pH 7.2) (see Recipes)
9. 50 mM Tris-HCl (pH 7.5) (see Recipes)
10. RNase A solution (see Recipes)
Recipes
1. 70% ethanol
Dissolve 70 mL of pure ethanol in 30 mL of deionized water. Store at 4 °C.
2. Normal saline
Dissolve 0.9 g of sodium chloride in 100 mL of deionized water, autoclave (121 °C, 20 min), and let cool down. Store at room temperature (RT).
3. 1.5 M NaCl solution
Dissolve 8.8 g of sodium chloride in approximately 80 mL of deionized water. After complete dissolution, the volume is adjusted to 100 mL with additional deionized water. Autoclave (121 °C, 20 min) and let cool down. Store at RT.
4. SYTOXTM green working solution
Dissolve 20 μL of SYTOXTM Green in 980 μL of DMSO. Store at -20 °C.
5. Percoll isotonic solution
100% Percoll, 90 mL
Autoclave (121 °C, 20 min) and let cool down. Store at RT.
Add 10 mL of 1.5 M NaCl solution into 90 mL of Percoll and vortex to mix.
Prepare Percoll isotonic solution just before separating. Long-term storage may cause Percoll particles to bind to salt ions due to gravity, which is not conducive to the generation of the density gradient.
6. YPDA culture medium
Yeast extract 1% (w/v), 10 g
Peptone A 2% (w/v), 20 g
Adenine 0.05% (w/v), 0.5 g
H2O, add up to 950 mL
Prepare solution, autoclave (121 °C, 20 min), and let cool down.
D-(+)-Glucose 2% (w/v), 20 g
H2O, add up to 50 mL
Prepare solution, autoclave (115 °C, 15 min), let cool down, and pour into the above solution.
For solid YPDA, add 1.5% agar (15 g/L).
7. Density marker beads
50 μL each of 1.02 g/L (green), 1.04 g/L (orange), 1.06 g/L (purple), 1.08 g/L (dark blue), 1.09 g/L (pink), and 1.13 g/L (light blue) density marker beads. Vortex to mix.
8. 50 mM sodium citrate buffer (pH 7.2)
Citric acid trisodium dihydrate 50 mM, 14.7 g
H2O, add up to 1,000 mL
Adjust the pH to 7.2 with citric acid
Prepare solution, autoclave (121 °C, 20 min), and let cool down. Store at RT.
9. 50 mM Tris-HCl (pH 7.5)
Tris 50 mM, 6.2 g
H2O, add up to 1,000 mL
Adjust the pH to 7.5 with hydrochloric acid.
Prepare solution, autoclave (121 °C, 20 min), and let cool down. Store at RT.
10. RNase A solution
1.5 M NaCl, 10 μL
50 mM Tris-HCl (pH 7.5), 200 μL
RNase A 10 mg/mL, 10 mg
Ethanol, 500 μL
ddH2O add up to 1 mL. Store at -20 ˚C
Equipment
1. Sterile pipette sets [Nantong Leierxin Experimental Equipment, catalog numbers: LP025 (1 mL), LP024 (200 μL), LP022 (10 μL)]
2. Centrifugal tube [Nantong Leierxin Experimental Equipment, catalog numbers: LP074 (2 mL), LP027 (1.5 mL)]
3. Plastic culture dishes, 85 mm × 15 mm (Nantong Leierxin Experimental Equipment)
4. Laminar airflow (Suzhou Antai Airtech, catalog number: SW-CJ-2FD)
5. Pipette [Eppendorf, catalog numbers: 4027959 (10 μL); 1224923 (20 μL); 4301409 (100 μL); 1599049 (200 μL); H39622F (1,000 μL); 236061z (5,000 μL)]
6. Heating magnetic stirrer (IKA works, catalog number: 07.012755)
7. Precision balance (Mettler Toledo, catalog numbers: AB204-N and PB602-N)
8. Centrifuge (Thermo Scientific, model: Pico 17 with 775003424 angle rotor)
9. Glass test tubes, 17 mm × 185 mm
10. Test tube caps, 21 mm
11. 250 mL Erlenmeyer flask
12. Icebox [Siemens, catalog number: BCD-254(KK25E73TI)]
13. Shaker incubator (GlobalFoundries, catalog number: 3031)
14. Light microscope (Nikon, catalog number: YS100)
15. Spectrophotometer (Pgeneral, catalog number: T6 Xinyue)
16. Biosensors analyzer (Sieman, catalog number: S-10)
17. Autoclave (Shanghai Boxun Medical Biological Instrument, catalog number: YXQ-50G)
18. Flow cytometer (Beckman Coulter, catalog number: CytoFLEX)
19. Flow cytometer (BD, model: BD Aria III)
20. Vortex mixer (Thermo Scientific, catalog number: 88880018)
21. Centrifuge (Hettich Zentrifugen, model: Universal 32R with 1689 angle rotor)
22. Centrifuge (Jouan CR422, catalog number: 11175330 with 11175338 M4 swing-out rotor)
23. Electric incubator (Shanghai Yiheng Scientific Instrument, catalog number: DHP 9082)
24. Laser confocal scanning microscope (LCSM) (Leica, model: Leica Stellaris 8 FALCON)
25. Sterilizing pot (Hirayama, catalog number: HV-85)
Software and datasets
1. FlowJo v10.8.1 https://www.flowjo.com/
2. LASX Office 1.4.7 28921 https://www.leica-microsystems.com/
Note: The above-mentioned software might not be freely accessible. Interested users should reach out to the sales departments of the corresponding companies for acquisition.
Procedure
文章信息
稿件历史记录
提交日期: Oct 2, 2024
接收日期: Mar 4, 2025
在线发布日期: Mar 31, 2025
出版日期: Apr 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Zu, C., Dong, J., Zhang, Y., Zhang, Y., Huang, Z. and Zou, S. (2025). A Miniaturized Percoll Gradient Method for Isolation of Quiescent Cells of Yeast. Bio-protocol 15(8): e5279. DOI: 10.21769/BioProtoc.5279.
分类
微生物学 > 体内实验模型 > 真菌
细胞生物学 > 细胞活力 > 细胞存活
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link