- EN - English
- CN - 中文
Rapid Plasmid-Free Generation of Recombinant Positive-Strand RNA Viruses That Use IRES-Mediated Translation Using an Expansion of the Circular Polymerase Extension Reaction (CPER)
基于扩展型CPER的快速无质粒重组正链RNA病毒构建方法:适用于IRES介导翻译系统
发布: 2025年04月20日第15卷第8期 DOI: 10.21769/BioProtoc.5275 浏览次数: 2083
评审: Emilie BesnardAnonymous reviewer(s)
Abstract
Reverse genetics systems in virology are technologies used to generate recombinant viruses, enabling the manipulation of viral genes. Recombinant viruses facilitate the investigation of pathogenesis and the development of antivirals. In studies of positive-sense single-stranded RNA (ssRNA) viruses, a reverse genetics approach typically uses infectious viral cDNA clones derived from bacterial artificial chromosomes and plasmids or from the in vitro ligation of viral cDNA fragments. However, these methods are time-consuming, involve complex procedures, and do not always successfully generate recombinant viruses. Possible reasons for unsuccessful outcomes include i) viral sequences exhibiting toxicity in bacterial systems, ii) the duplication of viral genes observed in some strains, complicating the acquisition of correct cDNA clones, and iii) certain cell lines being highly susceptible to infection but difficult to transfect with nucleotides. For these reasons, a simple and rapid reverse genetics system is needed to accelerate research on ssRNA viruses. The circular polymerase extension reaction (CPER) method offers a solution by eliminating the need for molecular cloning in bacteria, enabling the generation of recombinant viruses over a shorter timeframe. This method has been widely adopted for the study of ssRNA viruses, including SARS-CoV-2 and flaviviruses. Recently, we expanded the CPER method for ssRNA viruses using internal ribosome entry site (IRES)-mediated translation. This protocol details the experimental procedures, using bovine viral diarrhea virus as an example—one of the most challenging viruses for generating viral cDNA clones because of the factors listed above.
Key features
• Rapid generation of recombinant positive-strand RNA viruses.
• The CPER method eliminates the need for molecular cloning in bacteria, enabling the rapid generation of recombinant viruses.
• The CPER method for ssRNA viruses enables efficient translation of viruses using IRES by incorporating the gene cassette of RNA Pol-I promoters and terminators.
Keywords: Reverse genetics (反向遗传学)Graphical overview
Background
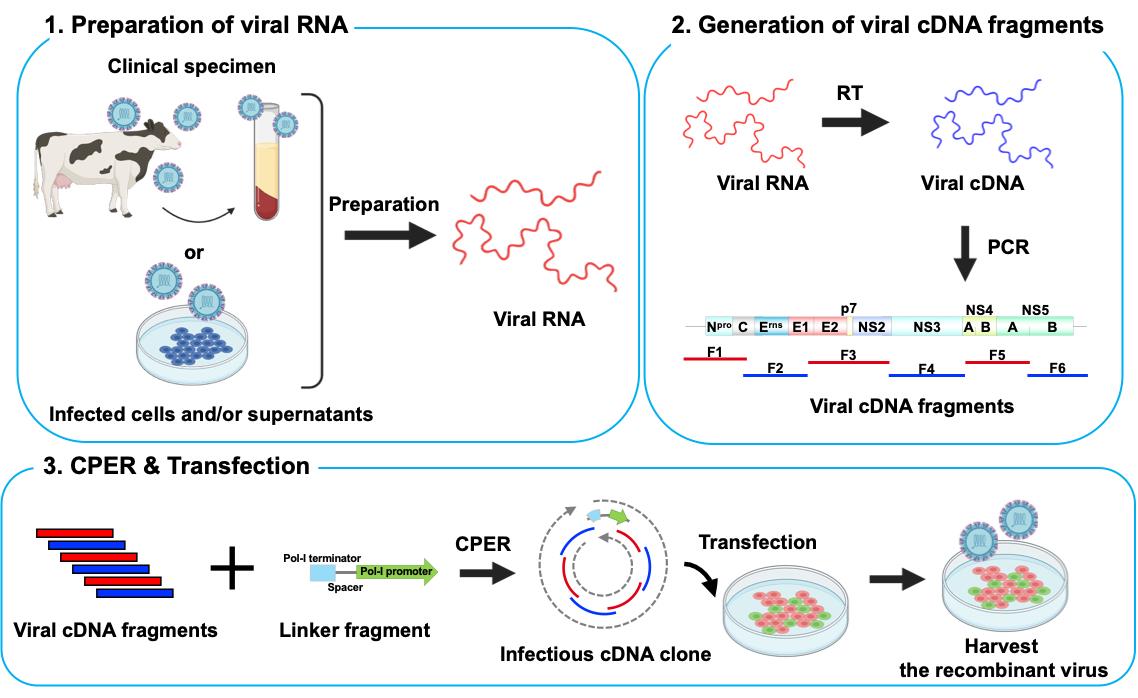
In virology, the reverse genetics system is a technology for generating recombinant viruses that enables the manipulation of viral genes. Using this system, we can define the function of viral genes/proteins and investigate the mechanisms of viral pathogenicity [1]. To date, infectious viral cDNA clones from bacterial artificial chromosomes and plasmids or from in vitro ligation of viral cDNA fragments have been used for reverse genetics in studies on positive-sense single-stranded RNA (ssRNA) viruses [2–5]. Although these methods allow for the generation of recombinant viruses, they are time-consuming and involve complicated procedures. Thus, a simple and efficient reverse genetics system is required, particularly in the context of pandemics caused by emerging/re-emerging ssRNA viruses.
A method for the generation of recombinant ssRNA viruses using circular polymerase extension reaction (CPER) has been developed [6,7]. In this method, full-length viral cDNA fragments with overlapping ends and an RNA Pol-II promoter-encoding linker are assembled into a circular genome by DNA polymerase and are directly transfected into cells to recover infectious viruses. This technology was applied to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which possesses one of the largest genomes (~30 kb) among the ssRNA viruses, enabling the rapid generation of recombinant viruses. It also enabled the rapid characterization of SARS-CoV-2 variants of concern and variants of interest after their emergence [8–12].
Flaviviruses and SARS-CoV-2 perform translation in a cap-dependent manner, whereas certain ssRNA viruses, such as hepatitis C virus (HCV) and bovine viral diarrhea virus (BVDV), perform cap-independent internal ribosome entry site (IRES)-mediated translation. The CPER method, optimized for efficient capping after viral RNA transcription, may not be directly applicable to ssRNA viruses that use IRES-mediated translation. Therefore, we adapted the CPER method using a gene cassette containing an RNA Pol-I promoter and terminator sequence that is needed for the efficient translation of RNA viruses [13].
BVDV, now renamed as Pestivirus bovis, belongs to the Pestivirus genus of the Flaviviridae family and possesses ssRNA that undergoes IRES-mediated translation. BVDV is the causative agent of bovine viral diarrhea, which leads to economic losses in agriculture. Generating recombinant BVDV using conventional reverse genetics has proven challenging for three reasons: i) the viral sequence exhibits toxicity in bacteria [14]; ii) gene duplication in some BVDV strains makes it difficult to obtain the correct cDNA clones [15]; and iii) MDBK cells, the most common cell line for BVDV propagation, show high susceptibility but are difficult to transfect with nucleotides [16].
In this protocol, we describe an optimized CPER method for ssRNA viruses using IRES-mediated translation, with BVDV serving as an example. Our method overcomes commonly encountered challenges and successfully produces recombinant viruses over a short timeframe. In addition, we demonstrate the generation of recombinant viruses without the need for isolation from permissive cell cultures, which is useful for the rapid characterization of viral strains, particularly during a pandemic.
Materials and reagents
Biological materials
1. Bovine MDBK cells (ATCC, catalog number: CCL-22)
2. Lenti-X 293T cells (TAKARA BIO, catalog number: 632180)
3. Serum from cattle persistently infected with BVDV (BVDV-1b strain Shihoro/B_6, Hirose et al. [17])
Reagents
1. BVDV antibody-free FBS (Japan Bio Serum, catalog number: 621-00535)
2. DMEM (4.5 g/L glucose) with L-glutamine, without sodium pyruvate, liquid (Nacalai Tesque, catalog number: 08459-64)
3. Penicillin-streptomycin mixed solution (Nacalai Tesque, catalog number: 26253-84)
4. PrimeSTAR® GXL DNA polymerase (TAKARA BIO, catalog number: 12292-04)
5. KOD One® PCR master mix (Toyobo, catalog number: 18538-01)
6. 1 kb DNA ladder (TAKARA BIO, catalog number: 3412A)
7. SuperScriptTM IV VILOTM master mix (Thermo Fisher Scientific, catalog number: 11756050)
8. Opti-MEM (Thermo Fisher Scientific, catalog number: 07088-54)
9. TransIT®-LT1 transfection reagent (Mirus, catalog number: MIR2304)
10. Acetone (Nacalai Tesque, catalog number: 00309-35)
11. D-PBS (-) without Ca and Mg, liquid (Nacalai Tesque, catalog number: 14249-24)
12. Anti-pestiviral NS3 antibody (prepared in-house, see Kameyama et al. [18])
13. Goat anti-mouse IgG Alexa Fluor 488-conjugated secondary antibody (Thermo Fisher Scientific, catalog number: A32723)
14. PureLinkTM RNA Mini kit (Thermo Fisher Scientific, catalog number: 12183018A)
15. FastGene Gel/PCR Extraction kit (NIPPON Genetics, catalog number: FG-91202)
16. Primer sets (Fasmac; Table 1)
Table 1. Primer sets used for CPER of BVDV-1b
| Fragment | Genome size (bp) | Orientation | Nucleotide sequence (5'–3') |
|---|---|---|---|
| BVDV-1b Fragment 1 | 1303 | Forward | GTATACGAGGTTAGGCAAGTTC |
| Reverse | GATTTTCTCTGGCCAGATC | ||
| BVDV-1b Fragment 2 | 2401 | Forward | GATCTGGCCAGAGAAAATC |
| Reverse | CTCTCTTAGTAGTAGGTATAGTAG | ||
| BVDV-1b Fragment 3 | 2316 | Forward | CTACTATACCTACTACTAAGAGAG |
| Reverse | CATGTATTGATAGACTGACTCAGCTGC | ||
| BVDV-1b Fragment 4 | 2501 | Forward | GCAGCTGAGTCAGTCTATCAATACATG |
| Reverse | CTTATCTTCCCTTCAGAGTCCATC | ||
| BVDV-1b Fragment 5 | 2236 | Forward | GATGGACTCTGAAGGGAAGATAAG |
| Reverse | GTGCTTCTCTGAGTCCAGTAC | ||
| BVDV-1b Fragment 6 | 1652 | Forward | GTACTGGACTCAGAGAAGCAC |
| Reverse | GGGGCTGTTAAGGGTTTTCCCTAGTC | ||
| BVDV-1b Linker fragment | 738 | Forward | ACCCTTAACAGCCCCCCCCCCCAACTTCGGAGGTCGAC |
| Reverse | GAACTTGCCTAACCTCGTATACAATAACCCGGCGGCCCAAAATGC |
Solutions
1. Maintenance medium (see Recipes)
2. 2% FBS medium (see Recipes)
Recipes
1. Maintenance medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM | Not applicable | 500 mL |
| BVDV antibody-free FBS | 9.1% | 50 mL |
| Penicillin-streptomycin | 1% | 5 mL |
2. 2% FBS medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM | Not applicable | 500 mL |
| BVDV antibody-free FBS | 2% | 10 mL |
| Penicillin-streptomycin | 1% | 5 mL |
Laboratory supplies
1. Collagen-coated microplate 6-well with lid, collagen type I (IWAKI, catalog number: 4810-010N)
2. Collagen-coated microplate 24-well with lid, collagen type I (IWAKI, catalog number: 4820-010)
3. Flat bottom micro tube 1.5 mL (BIO-BIK, catalog number: CF-0150)
4. 0.2 mL 8-strip PCR tube caps with dome top, natural (WATSON BIO LAB, catalog number:137-432C)
Equipment
1. CO2 incubator (PHCbi, model: MCO-170AICUVD)
2. NanoDrop Lite UV-Vis spectrophotometer ND-LITE (Thermo Fisher Scientific, catalog number: 32-1001)
3. Fluorescence microscope (KEYENCE, model: BZ-X810)
Software and datasets
1. Prism v. 10.2.2 (GraphPad, 7/31/2024)
Procedure
文章信息
稿件历史记录
提交日期: Jan 8, 2025
接收日期: Mar 11, 2025
在线发布日期: Mar 26, 2025
出版日期: Apr 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Yamamoto, H., Tamura, T. and Fukuhara, T. (2025). Rapid Plasmid-Free Generation of Recombinant Positive-Strand RNA Viruses That Use IRES-Mediated Translation Using an Expansion of the Circular Polymerase Extension Reaction (CPER). Bio-protocol 15(8): e5275. DOI: 10.21769/BioProtoc.5275.
分类
微生物学 > 微生物遗传学 > DNA > PCR
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link