- EN - English
- CN - 中文
Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
利用酰基树脂辅助捕获法(Acyl-RAC)检测与分析S-酰化蛋白
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5268 浏览次数: 1849
Abstract
Protein palmitoylation is a lipid modification where a palmitoyl group is covalently attached via a thioester linkage to one or more cysteines on a substrate protein. This modification, catalyzed by a group of enzymes named DHHC enzymes after their conserved Asp-His-His-Cys motif, plays a significant role in regulating the localization, stability, and function of a wide range of cellular and viral proteins. By influencing how and where proteins interact within the cell, palmitoylation is essential for various cellular processes, including signaling pathways, membrane dynamics, and protein–protein interactions. Here, we describe the acyl-RAC assay, a biochemical technique designed to specifically enrich and analyze palmitoylated proteins from complex biological samples, such as cell lysates or tissue extracts. The assay begins by reducing and blocking free cysteine thiol groups on proteins, ensuring that only those thiols involved in thioester bonds with palmitates are accessible for downstream analysis. These thioester bonds are then cleaved to release the fatty acids from the cysteines, which are subsequently captured using thiopropyl Sepharose beads that bind to the newly exposed thiol groups. The captured proteins are eluted from the beads by breaking the bond between the thiol and the resin with reducing agents, and the proteins are then analyzed by SDS-PAGE followed by western blotting to identify and quantify them. The acyl-RAC assay's specificity for S-palmitoylated proteins makes it an invaluable tool for exploring this modification. It not only allows for the identification of previously unknown palmitoylated proteins, thereby deepening our understanding of palmitoylation in cellular processes and viral infections, but it also enables quantitative comparisons of protein palmitoylation under different experimental conditions or treatments.
Key features
• Allows identification of acylated proteins.
• Quantitative analysis of S-palmitoylation levels under various conditions by western blot.
• Requires at least seven days to complete.
Keywords: SARS-CoV-2 (新冠病毒)Graphical overview
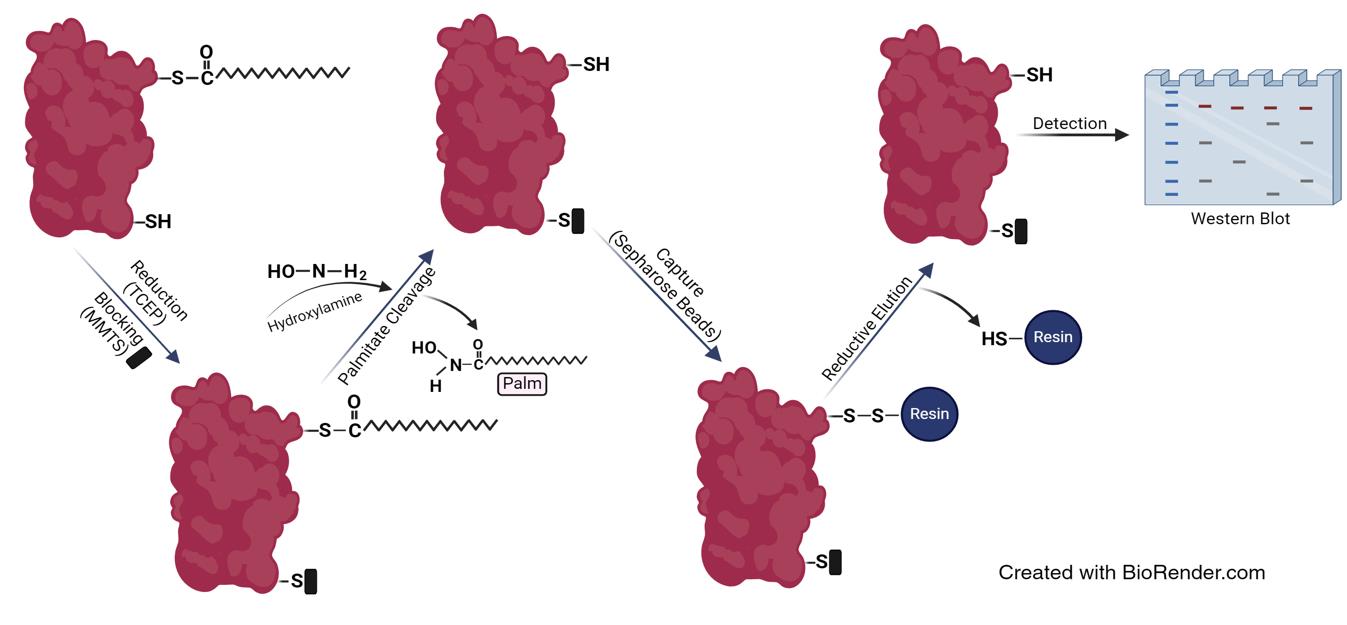
Background
Protein palmitoylation is a vital, reversible lipid modification that plays a key role in a wide range of cellular activities, such as membrane trafficking, cell signaling, and the formation of protein complexes [1]. This process extends beyond cellular proteins and also plays a significant role in the viral life cycle, aiding in the correct assembly and function of viral particles [2]. The study of protein palmitoylation can yield valuable insights into various diseases, including cancer, neurological disorders, and infectious diseases, where abnormal palmitoylation is frequently implicated [3]. Additionally, research in this field may contribute to the development of therapeutic strategies that target this critical modification [4].
The conventional and widely regarded "gold standard" approach for identifying acylated proteins is the utilization of radioactive palmitate, either labeled with [3H] or [14C], which incorporates into palmitoylation sites [5,6]. Nevertheless, this method is limited to studying S-acylation in cultured cells and cannot be employed for intact tissue. In addition to the safety concerns that arise due to radiation exposure, the technique's limited sensitivity in detecting low-abundance palmitoylated proteins and its low throughput make it less efficient for extensive studies [7]. Another approach for studying palmitoylation is a click chemistry–based method that involves labeling the substrate protein with an azide-linked fatty acid analog, 17-octadecynoic acid (17-ODYA), which attaches to palmitoylation sites and subsequently conjugates to a fluorescent tag with an azide group through a Cu (I)-catalyzed click chemistry reaction. SDS-PAGE is then used for in-gel fluorescence detection of the palmitoylated proteins using a Typhoon scanner. While this method is considered an effective tool for studying palmitoylation, it has several drawbacks, including the high cost of specialized tags and copper catalysts and inadequate sensitivity to detect low-abundance palmitoylated proteins [8,9].
The primary advantage of the acyl-RAC assay [10], along with the closely related acyl biotin exchange (ABE) assay [5], is their ability to detect palmitoylated proteins from both tissue and cultured cell samples. These assays utilize hydroxylamine to cleave thioester bonds, leveraging its nucleophilic properties to selectively target and break these bonds by attacking the carbonyl carbon. This specificity is due to the increased electrophilicity of thioesters, ensuring that amides, ester bonds, and non-palmitoylated cysteines remain intact. However, they cannot identify the specific type of bound fatty acid without mass spectrometry analysis. Additionally, if free cysteines are not adequately blocked, these methods may generate false palmitoylation signals. This issue can be mitigated by extending the reduction time and increasing the concentration of the blocking agent. The nonspecific detection of non-palmitoylated proteins can be investigated by processing an equal volume of the protein extract in parallel but without the hydroxylamine treatment step used to cleave thioester bonds. These negative control samples help confirm the protocol's specificity for detecting palmitoylated proteins [11].
Acyl-RAC has been employed to study the palmitoylation of various cellular and viral proteins [12,13]. In this protocol, we apply the acyl-RAC assay to investigate the role of certain amino acids of the SARS-CoV-2 spike protein in its palmitoylation, by comparing the palmitoylation levels of spike mutants with the wild type (WT). We selected the SARS-CoV-2 spike as a model since its palmitoylation is crucial for its membrane association, stability, and functionality [14,15]. Studying this modification provides valuable insights into this viral protein regulation and highlights potential therapeutic targets.
Materials and reagents
Biological materials
1. Human embryonic kidney (HEK) 293T cells (American Type Culture Collection, catalog number: ATCC CRL-11268)
2. pCAGGS plasmids with cloned codon-optimized sequence of WT or mutated spikes. The pCAGGS plasmid containing the WT spike sequence was kindly provided by Dr. Dusan Kunec, Free University of Berlin. Mutations in the spike were introduced using the QuickChangeTM site-directed mutagenesis protocol [16]
Reagents
Tissue culture
1. Dulbecco’s modified Eagle’s medium (DMEM) (PAN Biotech, catalog number: P04-05551)
2. Fetal bovine serum (FBS) (PAN Biotech, catalog number: P30-3306)
3. Penicillin/Streptomycin (10,000 U/mL) (PAN Biotech, catalog number: P06-07100)
4. Trypsin/EDTA (PAN Biotech, catalog number: P10-023100)
Transfection
1. Lipofectamine 3000 transfection reagent (Thermo Fisher, catalog number: L3000015)
2. Opti-MEMTM I reduced serum medium (Thermo Fisher, catalog number: 31985070)
Acyl-RAC
1. Thiopropyl agarose beads (Creative biomart, catalog number: Thio-001A)
2. Methyl methanethiosulfonate (MMTS) (Sigma-Aldrich, catalog number: 208795)
3. Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) (Carl Roth, catalog number: HN95.2)
4. cOmpleteTM, EDTA-free protease inhibitor cocktail (Roche, catalog number: 11873580001)
5. Sodium dodecyl sulfate (SDS) (Carl Roth, catalog number: 2326.4)
6. Acetone (Carl Roth, catalog number: 20T0.1)
7. Hydroxylamine hydrochloride (HA) (Thermo Fisher, catalog number: 159417-100G)
8. Tris (Carl Roth, catalog number: 2449.2)
9. Hydrochloric acid (HCL) (Carl Roth, catalog number: 2607.1)
10. Sodium hydroxide (NaOH) (Carl Roth, catalog number: 9356.1)
11. HEPES (Carl Roth, catalog number: HN77.3)
12. NaCl (Carl Roth, catalog number: 3957.1)
13. EDTA (AppliChem, catalog number: A2937)
14. Triton X-100 (Merck, catalog number: 8603)
SDS-PAGE and western blot
1. Tetramethylethylenediamine (TEMED) (Carl Roth, catalog number: 2367.3)
2. Ammonium persulfate (APS) (Carl Roth, catalog number: 9592.2)
3. Sodium dodecyl sulfate (SDS) (Carl Roth, catalog number: 2326.4)
4. Bromophenol blue (Sigma-Aldrich, catalog number: 1081220005)
5. 30% acrylamide/bisacrylamide (37.5:1) (Carl Roth, catalog number: 3029.1)
6. Anti SARS-CoV-2 spike (S2 subunit) mouse monoclonal antibody (GeneTex, catalog number: GTX632604)
7. HRP-conjugated anti-mouse secondary antibody (Bio-Rad, catalog number: 1706516)
8. Anti flotillin-2 polyclonal mouse antibody (BD Biosciences, catalog number: 610383)
9. Goat anti-mouse IgG (H + L) antibody with horseradish peroxidase conjugate (Bio-Rad, catalog number: 1706516)
10. Pierce ECL Plus reagent (Thermo Fisher, catalog number: 32132)
11. Skimmed milk powder (Carl Roth, catalog number: T145.3)
12. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
13. Tween 20 (Carl Roth, catalog number: 9127.1)
14. Glycerol (Carl Roth, catalog number: 3783.1)
15. 1,4-Dithiothreitol (DTT) (Carl Roth, catalog number: 6908.2)
16. KCl (Carl Roth, catalog number: HN02.3)
17. Na2HPO4 (Carl Roth, catalog number: 4984.4)
18. KH2PO4 (AppliChem, catalog number: A3095)
19. Methanol (Sigma-Aldrich, catalog number: 106009)
Solutions
1. Phosphate buffer saline (PBS) (see Recipes)
2. Phosphate buffer saline-Tween (PBST) (see Recipes)
3. Protease inhibitor 25× (see Recipes)
4. Buffer A (lysis buffer) (see Recipes)
5. Blocking buffer (see Recipes)
6. Binding buffer (see Recipes)
7. 5× reducing SDS-PAGE sample buffer (see Recipes)
8. Stacking SDS-PAGE gel solution (see Recipes)
9. Separating SDS-PAGE gel solution (see Recipes)
10. Membrane blocking solution (see Recipes)
11. 3% BSA (see Recipes)
12. Growth medium (see Recipes)
Recipes
Note: The shelf life of buffers stored at room temperature is up to six months. The shelf life of buffers stored at 4 °C or -20 °C is up to one year.
1. 1× Phosphate buffer saline (PBS)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| Na2HPO4 | 10 mM | 1.44 g |
| KH2PO4 | 1.8 mM | 0.24 g |
| dH2O | n/a | See note* |
| Total | n/a | 1 L |
*Note: Add water to 800 mL first, allow the solution to mix completely, and adjust the pH to 7.4 with HCl before topping up water to 1 L.
Autoclave and store at room temperature.
2. Phosphate buffer saline-Tween (PBST)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PBS | 1× | 999 mL |
| Tween-20 | 0.1% | 1 mL |
| Total | n/a | 1,000 mL |
Store at room temperature.
3. Protease inhibitor 25×
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| cOmpleteTM, EDTA-free protease inhibitor cocktail | n/a | 1 tablet |
| dH2O | n/a | 2 mL |
Store at -20 °C.
4. Buffer A (lysis buffer)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 25 mM | 5.96 g |
| NaCl | 25 mM | 1.46 g |
| EDTA | 1 mM | 0.37 g |
| Triton X-100 | 1.5% | 15 mL |
| dH2O | n/a | See note* |
| Protease inhibitor cocktail | 1× | Freshly added according to the volume needed of Buffer A |
| Total | n/a | 1 L |
*Note: Add water to 800 mL and allow the solution to mix completely before topping up water to 1 L.
Filter and store at 4 °C. pH should be 7.2–7.6.
5. Blocking buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 100 mM | 23.83 g |
| EDTA | 1 mM | 0.37 g |
| SDS | 87.5 mM | 25.23 g |
| dH2O | n/a | See note* |
| MMTS | 1.5% (v/v) | Freshly added according to the volume needed of blocking buffer |
| Total | n/a | 1,000 mL |
*Note: Add water to 800 mL and allow the solution to mix completely before topping up water to 1 L.
Filter and store at 4 °C. pH should be 7.2–7.6.
6. Binding buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 100 mM | 23.83 g |
| EDTA | 1 mM | 0.37 g |
| SDS | 35 mM | 10.09 g |
| dH2O | n/a | See note* |
| Total | n/a | 1,000 mL |
*Note: Add water to 800 mL first and allow the solution to mix completely before topping up water to 1 L.
Filter and store at 4 °C. pH should be 7.2–7.6.
7. 5× reducing SDS-PAGE sample buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris-HCL (pH 6.5) | 300 mM | 1.5 mL (from 1 M stock) |
| SDS | 10% (w/v) | 1 g |
| Bromophenol blue | 0.05% (w/v) | 5 mg |
| Glycerol | 50% (v/v) | 5 mL |
| 1,4-Dithiothreitol (DTT) | 500 mM | 0.77 g |
| dH2O | n/a | To 10 mL |
For long-term use, aliquot and store at -20 °C. Avoid repeated freeze-thaw cycles if possible.
8. Stacking SDS-PAGE gel solution
| Reagent | Final concentration | Quantity or Volume for 2 gels |
|---|---|---|
| 30% Acrylamide/Bis solution (37.5:1) | 5% (w/v) | 1.66 mL |
| 1.0 M Tris-HCl (pH 6.8) | 0.5 M | 1.26 mL |
| 10% SDS | 0.1% (w/v) | 100 μL |
| 10% APS | 0.1% (v/v) | 100 μL |
| TEMED | 0.1% (v/v) | 10 μL |
| dH2O | n/a | 7 mL |
Prepare fresh.
9. Separating SDS-PAGE gel solution
| Reagent | Final concentration | Quantity or Volume for 2 gels |
|---|---|---|
| 30% Acrylamide/bisacrylamide | 12% (w/v) | 8 mL |
| 1.5 M Tris-HCl (pH 8.8) | 1.5 M | 5 mL |
| 10% SDS | 0.2% (w/v) | 200 μL |
| 10% APS | 0.2% (v/v) | 200 μL |
| TEMED | 0.2% (v/v) | 20 μL |
| dH2O | n/a | 6.6 mL |
Prepare fresh.
10. Membrane blocking solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Skimmed milk powder | 3% (w/v) | 0.3 g |
| PBST | n/a | 10 mL |
Prepare fresh.
11. 3% BSA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| BSA | 3% (w/v) | 0.3 g |
| PBST | n/a | 10 mL |
Prepare fresh.
12. Growth medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DMEM | n/a | 445 mL |
| FBS | 10% | 50 mL |
| Penicillin/Streptomycin | 100 U/L | 5 mL |
Store at 4 °C.
Laboratory supplies
1. Sterile Pipette tips (10, 200, 1000 μL) (Sarstedt, catalog numbers: 70.3010.205, 70.3030.200, 70.3050.205)
2. Pipettes (5, 10, 25 mL) (Sarstedt, catalog numbers: 86.1253.001, 86.1254.001, 86.1685.001)
3. 6-well cell culture dishes (Sarstedt, catalog number: 83.3920.005)
4. T75 flasks (Sarstedt, catalog number: 83.3911.002)
5. Conical tubes (Sarstedt, catalog number: 62.554.002)
6. 1.5 mL microcentrifuge tubes (Sarstedt, catalog number: 72.706)
7. Polyvinylidene difluoride (PVDF) blotting membrane, 0.2 μm (VWR, catalog number: 732-3200)
8. Rotilabo® blotting papers (Carl Roth, catalog number: CL75.1)
Equipment
1. Pipette controller (Hirschmann Laboratories, model: Pipetus)
2. Sterile laminar flow (Bleymehl, model: ScanLaf)
3. HeracellTM 240i CO2 incubator, 240 L (Thermo Fisher, catalog number: 51032875)
4. Inverted microscope (Motic, model: AE20)
5. Tabletop centrifuge (Eppendorf, model: 5424R)
6. Biofuge Stratos centrifuge (Heraeus, catalog number: 11727)
7. Vortex Genie 2TM (Bender&Hobein AG, model: K-550 GE)
8. Orbital shaker (Peqlab, model: 0S-10)
9. -20 °C freezer (Liebherr, catalog number: 9005382246719)
10. Ice machine (Ziegra, model: 174059)
11. pH meter (Knick, model: 761-Calimatic)
12. Tube rotator (Carl Roth, model: ATX1.1)
13. Water bath shaker (Brunswick Scientific, model: C76)
14. SDS-PAGE system (Biometra, model: Mini-Gel Twin)
15. ThermoMixer® C heat block (Eppendorf, catalog number: 5382000023)
16. PowerPacTM HC power supply (Bio-Rad, model: 1645052)
17. CriterionTM vertical electrophoresis cell (Bio-Rad, model: 1656001)
18. CriterionTM empty cassettes (Bio-Rad, model: 3459901)
19. Chemiluminescence imaging system (Vilber Lourmat, model: Fusion SL)
20. Nanodrop (PeqLab, model: ND 1000)
Software and datasets
1. Image J v1.54f (NIH, 6/29/2023)
2. Prism v8.0.2 (GraphPad Software Inc., 6/2/2019)
3. Fusion v15.11 (Vilber Lourmat, 16/9/2019)
4. Microsoft Excel v16.54 (Microsoft corporation, 10/5/2021)
5. ND-1000 v3.0.7 (PeqLab, 2/17/2009)
Procedure
文章信息
稿件历史记录
提交日期: Oct 31, 2024
接收日期: Mar 4, 2025
在线发布日期: Mar 23, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Abdulrahman, D. A. and Veit, M. (2025). Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC). Bio-protocol 15(7): e5268. DOI: 10.21769/BioProtoc.5268.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 修饰
生物化学 > 蛋白质 > 翻译后修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













