- EN - English
- CN - 中文
Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
贴壁细胞、悬浮细胞及肿瘤组织中位点特异性同源重组活性的检测方法
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5260 浏览次数: 2446
评审: Ivan ShapovalovAnonymous reviewer(s)
Abstract
Homologous recombination (HR) is a major pathway to repair DNA double-strand breaks. Hereditary breast and ovarian cancer syndrome (HBOC) is caused by germline pathogenic variants of HR-related genes, such as BRCA1 and BRCA2 (BRCA1/2). Cancer cells with HR deficiency are sensitive to poly(ADP-ribose) polymerase (PARP) inhibitors. Therefore, accurate evaluation of HR activity is helpful to diagnose HBOC and predict the effects of PARP inhibitors. The direct-repeat GFP (DR-GFP) assay has been utilized to evaluate cellular HR activity. However, evaluation by the DR-GFP assay tends to be qualitative and requires the establishment of stable cell lines. Therefore, we developed an assay to quantitatively measure HR activity called Assay for Site-Specific HR Activity (ASHRA), which can be performed by transiently transfecting two plasmids. In ASHRA, we use Cas9 endonuclease to create DNA double-strand breaks at specific sites in the genome, enabling the targeting of any endogenous loci. Quantification of HR products by real-time PCR using genomic DNA allows HR activity evaluated at the DNA level. Thus, ASHRA is an easy and quantitative method to evaluate HR activity at any genomic locus in various samples. Here, we present the protocols for adherent cells, suspension cells, and tumor tissues.
Key features
• This assay quantitatively evaluates homologous recombination (HR) activity.
• This assay can measure HR activity in adherent cells, suspension cells, and tumor tissues.
• This real-time PCR-based assay does not require a flow cytometer or next-generation sequencer.
Keywords: Homologous recombination (同源重组)Graphical overview
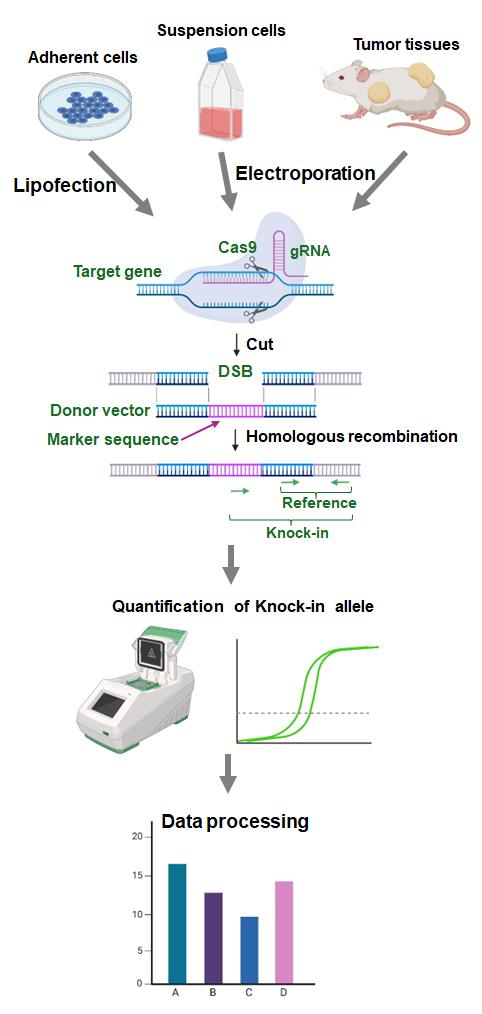
Background
Homologous recombination (HR) is an essential pathway to repair DNA double-strand breaks (DSBs) and contributes to the repair of interstrand crosslinks. Germline pathogenic variants of HR-related genes, such as BRCA1 and BRCA2 (BRCA1/2), cause hereditary breast and ovarian cancer syndrome (HBOC) [1]. Sporadic cancers also have alterations of HR-related genes [2]. Cancer cells with HR deficiency are sensitive to cancer treatments, including poly(ADP-ribose) polymerase (PARP) inhibitors, DNA-damaging agents such as platinum agents, and ionizing radiation [3,4]. Thus, accurate evaluation of HR activity is helpful to diagnose HBOC and stratify patients for cancer therapy.
For diagnosis of HBOC and stratification of patients for treatment with PARP inhibitors, genetic tests are performed to detect pathogenic variants of BRCA1/2 and other HR-related genes in the clinic. However, variants of uncertain significance have been identified, and an accurate functional assay to evaluate HR activity is required for appropriate clinical decision-making [5]. In addition, genomic scarring assays are used to detect HR deficiency in tumor tissues and predict the effects of PARP inhibitors [6] but do not detect changes in HR activity caused by revertant mutations of HR-related genes.
To evaluate cellular HR activity, the direct-repeat GFP (DR-GFP) assay has been utilized [7,8]. However, this assay targets the exogenous GFP locus and detects HR products at the protein level to evaluate HR activity, and its results tend to be qualitative [9]. In addition, the DR-GFP assay requires the establishment of a stable cell line [7,8]. DSB repair can be evaluated by immunostaining for RAD51 (an essential HR factor) or g-H2AX (a DSB marker). However, evaluation of RAD51 foci does not reflect HR activity due to alteration of steps downstream of RAD51 focus formation, and DSBs detected by g-H2AX foci are repaired by non-homologous end-joining in addition to HR [10,11]
To overcome these limitations, we developed an assay to evaluate HR activity called Assay for Site-Specific HR Activity (ASHRA) (Figure 1) [12]. In ASHRA, we use Cas9 endonuclease to create DSBs at specific sites in the genome, enabling the targeting of any endogenous loci. The marker sequence in the donor vector is integrated into the genome during the repair of the DSB by HR. The efficiency of integration of the marker sequence is evaluated by real-time PCR.
Quantification of HR products at the DNA level allows the method to be quantitative [9]. Furthermore, ASHRA can be performed by transiently transfecting two plasmids and does not require the establishment of a stable cell line. We successfully evaluated HR activity in lymphocyte-derived cells and tumor tissues by ASHRA using electroporation for plasmid delivery [13]. The direct measurement of HR activity in tumor tissues accurately predicted sensitivity to PARP inhibitors in vivo [13]. Thus, ASHRA can be performed using a wide range of samples, which greatly widens the application of HR activity evaluation.
Collectively, ASHRA allows easy and quantitative evaluation of HR activity at any locus of interest in the genome. Using ASHRA, we can directly evaluate HR activity in various samples irrespective of the causal genes, type of sequence variations, and shortage of functional information about the variation. In this article, we present the protocols of ASHRA for the measurement of HR activity in adherent cells, suspension cells, and tumor tissues.
Materials and reagents
Biological materials
1. Plasmids for ASHRA [LentiCRISPRv2-ACTB-C1 (Addgene: #169796), LentiCRISPRv2-scr (Addgene: #169795), and pBS-ACTB-C200-GFPfr1 (Addgene: #169798)] (Figure 1) (see General notes for additional information about plasmids)
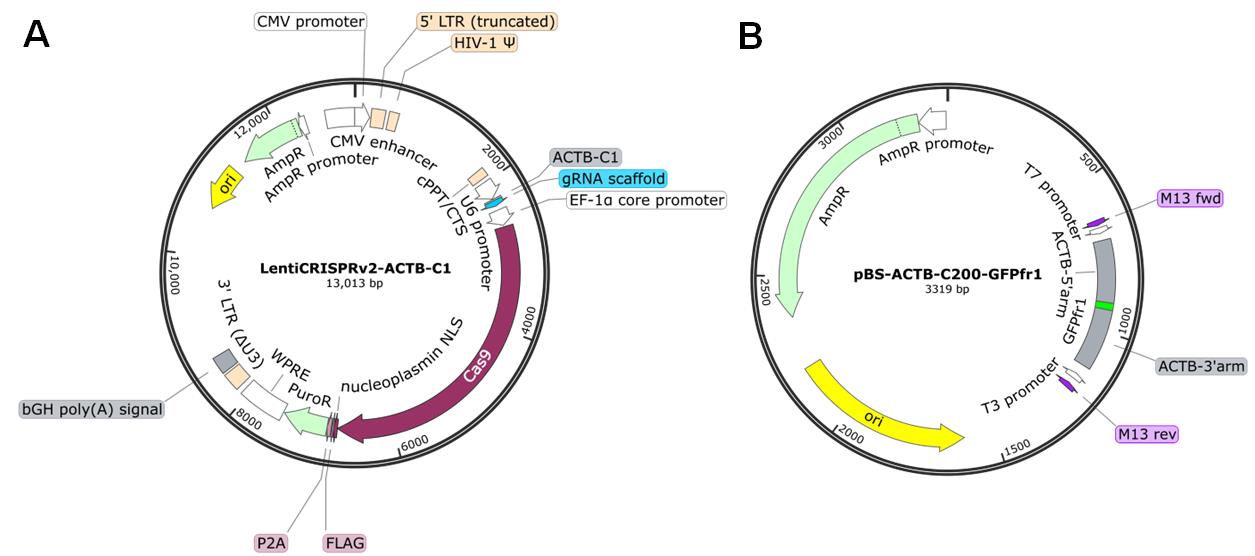
Figure 1. Maps of the plasmids. (A) LentiCRISPRv2-ACTB-C1 plasmid. This plasmid simultaneously expresses gRNA and Cas9 in transfected cells. LentiCRISPRv2-scr has a scrambled sequence instead of the ACTB-C1 gRNA sequence. (B) pBS-ACTB-C200-GFPfr1 plasmid. The priming sequence (GFPfr1) flanked by the donor arms for HR is cloned into the multiple cloning sites of pBlueScript SK II. Plasmid maps were generated with SnapGene software.
2. Primers, qPCR grade (Table 1) (see General notes for additional information about primers)
Table 1. Primer sets used to detect the knock-in allele (HR product) and reference allele. Sequences are shown in the 5'–3' direction.
| Forward primer | Reverse primer | |
|---|---|---|
| Knock-in allele | GTCCTGCTGGAGTTCGTGACCG | GTGCAATCAAAGTCCTCGGC |
| Reference allele | AGTTGCGTTACACCCTTTCTTG |
Reagents
1. TransIT-X2 dynamic delivery system (Mirus Bio, catalog number: MIR 6000)
2. PEI MAX (Polysciences, catalog number: 24765)
3. Opti-MEM (Thermo Fisher Scientific, catalog number: 11058021)
4. Culture medium and dishes appropriate for the cells used
5. FavorPrep Blood Genomic DNA Extraction Mini kit (Favorgen, catalog number: FABGK 001) or FavorPrep Tissue Genomic DNA Extraction Mini kit (Favorgen, catalog number: FATGK 001)
6. GoTaq qPCR master mix (Promega, catalog number: A6001)
7. NaOH (FUJIFILM Wako, catalog number: 194-18865)
8. Glucose (FUJIFILM Wako, catalog number: 049-31165)
9. Indigo carmine (FUJIFILM Wako, catalog number: 090-00082)
10. FBS (BioWest)
Solutions
1. PEI Max solution (see Recipes)
2. Real-time PCR mixture (see Recipes)
Recipes
1. PEI Max solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PEI MAX | 5 mg/mL | 100 mg |
| 1 N NaOH | n/a | Adjust pH to 7.6 |
| H2O | n/a | Up to 20 mL |
| Total | n/a | 20 mL |
2. Real-time PCR mixture
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| GoTaq qPCR master mix | 1× | 10 μL |
| Forward primer (10 μM) | 200 nM | 0.4 μL |
| Reverse primer (10 μM) | 200 nM | 0.4 μL |
| Template DNA | 10–50 ng/reaction | 1–5 μL |
| H2O | n/a | Up to 20 μL |
| Total | n/a | 20 μL |
Laboratory supplies
1. Electroporation cuvettes with 2 mm gap (NEPA Gene, catalog number: EC-002S)
2. 1 mL syringe with a 29G needle (Terumo, catalog number: SS-010F2913T)
3. Hard-shell 96-well PCR plates, low profile, thin wall, skirted (Bio-Rad, catalog number: HSP9601)
4. Microseal B (Bio-Rad, catalog number: MSB1001)
Equipment
1. CO2 incubator for cell culture (any type)
2. Electroporator (NEPA Gene, model: NEPA21 type2)
3. Forceps-type electrode (NEPA Gene, model: CUY650P7)
4. Thermal cycler for real-time PCR (Bio-Rad, model: CFX96)
Procedure
文章信息
稿件历史记录
提交日期: Dec 4, 2024
接收日期: Feb 23, 2025
在线发布日期: Mar 14, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Yoshino, Y., Kikuta, S. and Chiba, N. (2025). Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues. Bio-protocol 15(7): e5260. DOI: 10.21769/BioProtoc.5260.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













