- EN - English
- CN - 中文
Analysis of Modified Plant Metabolites Using Widely Targeted Metabolite Modificomics
基于广泛靶向代谢修饰组学的植物修饰代谢产物分析
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5259 浏览次数: 1468
评审: Tasleem JavaidAnonymous reviewer(s)
Abstract
Metabolite modifications play a critical role in enhancing plants’ adaptability to environmental changes and serve as a major source of functional diversity in metabolites. However, current metabolomics approaches are limited to targeted analyses of a small number of known modified metabolites and lack comprehensive, large-scale studies of plant metabolite modifications. Here, we describe a widely targeted metabolite modificomics (WTMM) strategy, developed using ultra-high-performance liquid chromatography–quadrupole linear ion trap (UHPLC-Q-Trap) and ultra-high-performance liquid chromatography–Q-Exactive Orbitrap (UHPLC-QE-Orbitrap) technologies. This strategy enables high-throughput identification and sensitive quantification of modified metabolites. Using tomato as a model, we conducted a metabolite modificomics study and constructed a WTMM database, identifying 165 novel modified metabolites. The WTMM strategy is broadly applicable and can be extended to the study of other plant species.
Key features
• WTMM enables large-scale detection and quantitative analysis of plant-modified metabolites.
• Integration of UHPLC-Q-Trap and UHPLC-QE-Orbitrap technologies.
• The WTMM database is extensible and applicable to other plant species.
Keywords: Metabolomics (代谢组学)Graphical overview
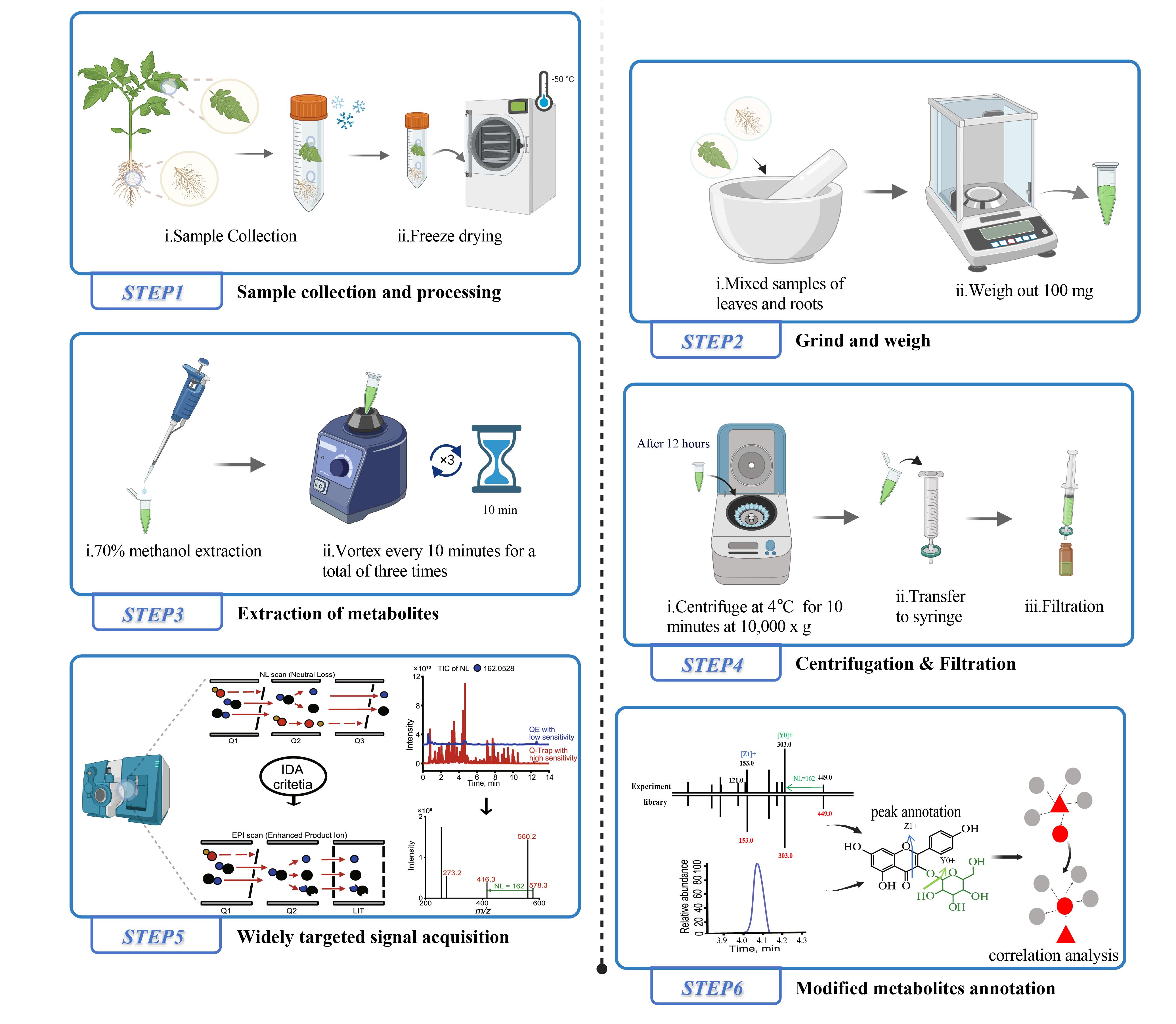
Brief description of the tomato tissue extraction process for liquid chromatography–mass spectrometry (LC–MS) analysis and the widely targeted metabolite modificomics (WTMM) strategy. The WTMM strategy employs an ultra-high-performance liquid chromatography–quadrupole-linear ion trap (UHPLC-Q-Trap) system in a stepwise neutral loss-enhanced product ion (NL-EPI) scanning mode to achieve high-throughput acquisition of modified metabolite profiles. Concurrently, high-resolution mass spectrometry data collected by the UHPLC-Q-Exactive-Orbitrap (UHPLC-QE-Orbitrap) system are utilized for structural annotation of metabolic signals. By applying the recursive algorithm of MetDNA, unknown metabolites (gray) are inferred based on correlation analysis with already identified metabolites (red). Created with BioRender.com.
Background
Structural modifications of metabolites in plants play a pivotal role in various biological processes, including growth and development [1,2]. Different types of structural modifications endow metabolites with distinct chemical properties. For example, immature tomato fruits contain the toxic substance α-tomatine, which provides protection against predation. During fruit ripening, α-tomatine undergoes further acylation and glycosylation to produce the non-toxic substance esculeoside A, thereby enhancing the nutritional quality of the fruit [3]. These diverse types of modified metabolites constitute the metabolite modificomics, offering valuable insights into the diversity of plant metabolites.
There are many unknown types of structural modifications in plant metabolites, and current metabolomics techniques for detecting modified metabolites each have their own limitations and advantages. Nontargeted approaches allow for large-scale detection of metabolite signals but often lack the sensitivity required to detect ultra-trace levels of unknown modified metabolites [4]. Conversely, targeted methods are highly sensitive but can only detect known modifications, making it difficult to systematically study the full range of chemical modifications in plants [5]. The strategy presented in this study integrates the strengths of high sensitivity, broad coverage, and high resolution to enhance the detection sensitivity and identification efficiency for modified metabolites.
Here, we describe a widely targeted metabolite modificomics (WTMM) strategy [6], which employs the stepwise neutral loss-enhanced product ion (NL-IDA-EPI) method using ultra-high-performance liquid chromatography–quadrupole linear ion trap (UHPLC-Q-Trap) to capture metabolic signals containing various modified groups. High-resolution mass spectrometry (HRMS) data collected via ultra-high-performance liquid chromatography-Q-Exactive Orbitrap (UHPLC-QE-Orbitrap) are then used, alongside metabolite databases, for structural annotation of these signals, enabling the construction of a WTMM database. Furthermore, we utilize the scheduled multiple reaction monitoring (sMRM) mode of Q-Trap to perform quantitative analysis of the signals in the WTMM database, optimizing detection parameters to improve data quality. Using tomato as a model, we applied the WTMM strategy to establish a tomato-specific WTMM database, identifying 165 novel modified metabolites. Additionally, we demonstrated the utility of this database by validating diagnostic biomarkers for bacterial wilt in tomatoes [6].
Materials and reagents
Biological materials
1. Tomato (MicroTom) seeds were surface sterilized at 50 °C for 30 min, germinated on moist filter paper in Petri dishes for two days, and then sown into trays filled with nursery soil. After five weeks, seedlings were transplanted into pots awaiting sampling or further treatment and planted at a density of 16.5 cm between plants in a row and 26 cm apart. Greenhouse management, including irrigation, fertilization, and pest control, essentially followed normal agricultural practices.
Reagents
1. Lidocaine (Sigma-Aldrich, catalog number: L1026)
2. Methanol (HPLC grade) (Merck, catalog number: 1.06035.2500)
3. Acetic acid (HPLC grade) (Merck, catalog number: 5.43808.0100)
4. Deionized water (Thermo Fisher Scientific, Lab Tower EDI 15 system)
5. Acetonitrile (HPLC grade) (Merck, catalog number: 1.00029.2500)
Solutions
1. 70% methanol extraction solvents (see Recipes)
2. Liquid chromatography solvent A (see Recipes)
3. Liquid chromatography solvent B (see Recipes)
Recipes
1. 70% methanol extraction solvents (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | n/a | 700 mL |
| Deionized water | n/a | 300 mL |
| Lidocaine | 1 mg/L | 1 mg |
Note: Store at 4 °C.
2. Liquid chromatography solvent A (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetic acid | n/a | 0.4 mL |
| Deionized water | n/a | 1,000 mL |
Note: Prepare before use.
3. Liquid chromatography solvent B (1,000 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetic acid | n/a | 0.4 mL |
| Acetonitrile | n/a | 1,000 mL |
Note: Prepare before use.
Laboratory supplies
1. 150 μL glass inserts (Agilent, catalog number: 5183-2088)
2. 2 mL screw-top vials and caps (Agilent, catalog number: 5182-0716)
3. Captiva syringe filters, ValueLab 13 mm, 0.2 μm (Agilent, catalog number: 5190-5269)
4. 2 mL plastic centrifuge tube (Labshark, catalog number: 130201014)
5. Shim-pack GISS C18 column 2 × 150 mm, 5 μm (Shimadzu, catalog number: 227-30066-03)
6. 2 mL disposable syringes (Agilent, catalog number: 5610-2111)
Equipment
1. Shimadzu Nexera X2 UHPLC system (Shimadzu, model: Nexera X2)
2. LC-MS/MS system (AB SCIEX, model: QTRAP 6500)
3. LC-MS/MS system (Thermo Fisher Scientific, model: Q Exactive Plus Orbitrap)
4. Mixer mill (Retsch, model: MM 400)
5. 1730R high-speed refrigerated microcentrifuge (Genespeed, catalog number: GS-1730R-NA)
6. MS 3 BASIC vortex oscillator (IKA, catalog number: 0003617000)
7. Freeze dryer (Christ, model: ALPHA 2-4 LD plus)
8. Ultra-low freezer, -80 °C (Sanyo, catalog number: MDF-U35V)
Software and datasets
1. Analyst 1.6.3 (AB SCIEX, https://sciex.com)
2. Compound Discoverer 3.1 (Thermo Fisher Scientific)
3. MS2 Analyzer (http://fiehnlab.ucdavis.edu/projects/MS2Analyzer/)
4. MetDNA (http://metdna.zhulab.cn/)
5. Multi Quant 3.0.3 (AB SCIEX)
Procedure
文章信息
稿件历史记录
提交日期: Dec 30, 2024
接收日期: Mar 4, 2025
在线发布日期: Mar 13, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Zhang, J., Li, S., Han, Y., Wang, S., Liu, P. and Yang, J. (2025). Analysis of Modified Plant Metabolites Using Widely Targeted Metabolite Modificomics. Bio-protocol 15(7): e5259. DOI: 10.21769/BioProtoc.5259.
分类
植物科学 > 植物新陈代谢 > 代谢物谱
系统生物学 > 代谢组学 > 全生物体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













