- EN - English
- CN - 中文
Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
甜樱桃发育中花原基的淀粉酶法定量检测
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5256 浏览次数: 1868
评审: Demosthenis ChronisAnonymous reviewer(s)
Abstract
Starch is a carbohydrate widely used in the plant kingdom as a fuel for different physiological processes. While different techniques are available for the quantification of starch stored in seeds and bark tissues, they have hardly been used to quantify starch content in developing flower buds, where starch has been reported to accumulate in different reproductive organs. Here, we detail a quantitative enzymatic method to measure starch concentration in developing flower primordia in sweet cherry (Prunus avium L.). First, starch is enzymatically hydrolyzed to D-glucose, which was then quantified by an enzyme-coupled assay involving hexokinase (HK) and glucose-6-phosphate dehydrogenase (G6PD) and spectrophotometric quantification of NADH absorbance at 340 nm. This method is a sensitive, rapid, and affordable protocol specifically optimized for tiny flower buds with low starch content. The technique is revealed to successfully determine starch content in non-freshly harvested samples—frozen and stored at -20 °C or stored in fixatives—allowing a temporal separation of sampling and quantification and making the protocol suitable for high-throughput experimental designs in different fields of plant research.
Key features
• Sensitive protocol to quantify starch content in developing flower primordia of sweet cherry.
• Starch quantification based on enzymatic hydrolysis of starch and spectrophotometric quantification of NADH absorbance at 340 nm using the hexokinase and glucose-6-phosphate dehydrogenase coupled assay.
• Accurately detects starch in very small samples with low starch concentrations in both fixed and frozen plant material.
Keywords: Starch enzymatic assay (淀粉酶法检测)Background
Starch is a carbohydrate widely produced in plants and composed of homopolymers of glucose, consisting mainly of two glucans, amylopectin and amylose [1]. In woody plant species, starch is the main storage carbohydrate [2], which accumulates in dedicated plant organs, covering the long-term glucan needs that fuel different processes such as germination or regrowth after certain periods of dormancy [3]. In addition, starch sustains the development of different plant reproductive tissues [4]. Various waves of amylogenesis and amylolysis have been reported during stamen and anther development [4–7] and during pollen tube growth within the style and the ovary [8,9]. Although the physiological and developmental functions of starch accumulation in reproductive organs are not fully understood, it has been suggested to support the development of reproductive organs and the male and female germline during flower development, the heterotopic growth of pollen tubes during the progamic phase, and the development of the embryo and seed after fertilization [4,8,10] and underpin the likelihood of a flower to become a fruit [11,12].
Different methodologies have been used to quantify starch content, such as high-performance liquid chromatography, gas chromatography, hydrolysis with perchloric or sulfuric acids, and enzymatic methods ([13] and references therein). Although these methods have been applied on leaves, roots, shoots, or bark tissues, they have not been used in developing flower buds harboring one to several tiny flower primordia, probably due to their small size and low starch concentration. Instead, starch quantification in these small reproductive structures has been performed using histochemical microscopy techniques coupled with a specific image analysis system, which can only provide relative values of starch content [14,15]. The enzymatic hydrolysis of starch coupled with the spectrophotometric assay of hexokinase activity, originally used to quantify starch content in fresh leaves [16,17] and later optimized for fresh small flower organs of Arabidopsis [4], has been further optimized in this work to quantify starch content in small developing reproductive organs of cultivated woody plant species like sweet cherry (Prunus avium L.).
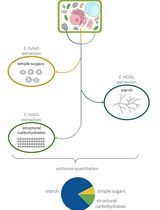
Our main objective was to optimize the enzymatic methodology to quantify starch concentration in small developing flower primordia with low starch concentrations. For this purpose, starch was gelatinized and enzymatically digested by amylase and amyloglucosidase into D-glucose. Next, a coupled hexokinase/glucose-6-phosphate dehydrogenase (HK/G6PD) enzyme assay was used to convert D-glucose into a stoichiometric amount of gluconate-6-phosphate and NADH. NADH was finally measured using a microplate reader, as it absorbs UV light at 340 nm. To make it useful for plant research applications where samples are routinely processed for histochemistry, this method is optimized for samples stored in fixative or frozen at -20 °C. This alternative enables developmental time-series sampling and long-term preservation for high-throughput sample processing and measurements. Thus, this protocol has the advantages of being simple, sensitive, and quick, not using harmful reagents, requiring reasonable labor, and not requiring specific laboratory equipment other than a UV spectrophotometer. The protocol has proven to be sensitive to very low sample weights (less than 1 mg) and low starch concentration (less than 0.18 mg/g of frozen or fixed sample), and the same results were obtained with both frozen and fixed samples.
The development of this enzymatic approach, applied for the first time to a fruit species, enables the quantification of starch in very small samples, such as reproductive structures. Previously, starch in these structures could only be assessed through histochemistry combined with image analysis, which allowed for the determination of relative content but not total quantification. Additionally, this method is equally effective with both frozen and fixed material, significantly extending the storage time of the samples.
Materials and reagents
Plant material
1. Flower buds from sweet cherry (Prunus avium L.) cultivar Earlise
a. Preserved in Carnoy’s fixative [samples are submerged into 96% ETOH/acetic acid glacial (3:1) for 24 h and then stored in 80% ETOH at 4 °C]
b. Preserved directly frozen at -20 °C
Reagents
1. 80% and 96% (v/v) ethanol (ETOH) (PanReac, catalog number: 141085)
2. Distilled water (dH2O) (Ecomatic, Wasserlab)
3. Ultrapure water (Ultramatic, Wasserlab)
4. Acetic acid glacial (Sigma, catalog number: 695092-2.5L)
5. Sodium acetate anhydrous (Sigma, catalog number: W302406)
6. Amyloglucosidase (Roche, catalog number: 11202332001)
7. Alpha-amylase (Roche, catalog number: 11202332001)
8. 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (Sigma, catalog number: H3375)
9. Hexokinase (Roche, catalog number: 11426362001)
10. ATP (Merck, catalog number: A2383-1G)
11. NAD+ (Roche, catalog number: 10127965001)
12. MgCl2 (Sigma, catalog number: M8266-100G)
13. Glucose 6-phosphate dehydrogenase (G6PD) (Roche, catalog number: 10165875001)
14. D-glucose (Sigma, catalog number: G8273-100G)
15. Lugol’s iodine solution (Sigma, catalog number: 32922)
16. Sodium hydroxide (NaOH) (Sigma, catalog number: S5881)
17. Hydrochloric acid (HCl) (Sigma, catalog number: 258148)
Solutions
1. 200 mM sodium acetate buffer (pH 4.8) (see Recipes)
2. 100 mM HEPES (pH 7.5) (see Recipes)
3. 100 mM ATP (see Recipes)
4. 100 mM NAD+ (see Recipes)
5. 100 mM MgCl2 (see Recipes)
6. Digestion mix (see Recipes)
7. Glucose assay mix (see Recipes)
8. Glucose stock solution 2.5 mM (see Recipes)
Recipes
1. 200 mM sodium acetate buffer (pH 4.8)
a. Solution A: Acetic acid glacial (MW 60.05 g/mol, d= 1.049 g/mL) 200 mM: 1.15 g in 100 mL of dH2O.
b. Solution B: Sodium acetate anhydrous (MW 82.03 g/mol) 200 mM: 1.64 g in 100 mL of dH2O.
c. Mix 20 mL of solution A, 30 mL of solution B, and 50 mL of dH2O.
2. 100 mM HEPES (pH 7.5)
a. Add 2.383 g of HEPES (MW 238.3 g/mol) to 80 mL of dH2O.
b. Adjust pH to ~7.5 using NaOH and HCl.
c. Complete with dH2O to 100 mL.
3. 100 mM ATP
Add 100 mg of ATP to 1.81 mL of ultrapure H2O.
4. 100 mM NAD+
Add 1 g of NAD+ to 15 mL of dH2O.
5. 100 mM MgCl2
Add 9.521 g of MgCl2 to 1,000 mL of dH2O.
6. Digestion mix
200 mM sodium acetate buffer, amyloglucosidase (6.3 U/mL), and alpha-amylase (50 U/mL). Example for 6 mL (40 samples × 150 µL each):
| Reagent | Quantity/Volume |
|---|---|
| Amyloglucosidase | 6.3 mg |
| Alpha-amylase | 30 µL |
| Sodium acetate buffer | 5,963.7 µL |
7. Glucose assay mix
100 mM HEPES pH 7.5, 1 mM MgCl2, 1 mM ATP, 1 mM NAD+, and hexokinase (6 U/mL). Example for 15 mL (96 samples × 150 µL each):
Note: Prepare extra solution when using a multipipette as it discards at least two dispensing volumes.
| Reagent | Volume |
|---|---|
| MgCl2 | 150 µL |
| ATP | 150 µL |
| NAD+ | 150 µL |
| Hexokinase | 60 µL |
| HEPES | 14,490 µL |
8. Glucose stock solution 2.5 mM
Add 0.045 g of D-glucose to 100 mL of dH2O.
Laboratory supplies
1. Microtubes 0.6 and 1.5 mL
2. Screw-capped microtubes 1.5 mL
3. Pipette tips (2–20, 20–200, and 100–1,000 µL)
4. Combi tips (0.1 and 10 mL)
5. 96-well microplates (Greiner, UV-STAR, catalog number: 655801)
Equipment
1. Thermoblock (Daihan Scientific, model: MaXtable H20)
2. Microcentrifuge (Beckman Coulter, model: Microfuge 16)
3. Analytical balance (Sartorius, model: Secura225-1S)
4. Pipettes (Gilson) (2–20, 20–200, and 100–1,000 µL)
5. Multipipette (Eppendorf, model: Multipette E3)
6. Incubator (Memment GmbH & Co., model: IN11)
7. Microtiter plate reader (Agilent, model: BioTek Epoch)
8. TissueLyser (Qiagen, model: TissueLyser II)
Procedure
文章信息
稿件历史记录
提交日期: Nov 22, 2024
接收日期: Feb 20, 2025
在线发布日期: Mar 12, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Santolaria, N., Fadón, E., Rodrigo, J. and Hedhly, A. (2025). Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry. Bio-protocol 15(7): e5256. DOI: 10.21769/BioProtoc.5256.
分类
植物科学 > 植物生理学 > 植物生长
植物科学 > 植物生物化学 > 糖类
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link