- EN - English
- CN - 中文
Screening for Streptococcus agalactiae: Development of an Automated qPCR-Based Laboratory-Developed Test Using Panther Fusion® Open AccessTM
链球菌筛查:基于Panther Fusion® Open Access™平台开发的自动化qPCR实验室自建检测方法
发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5255 浏览次数: 2082
评审: Satya Ranjan SahuAnonymous reviewer(s)
Abstract
Laboratory-developed tests (LDTs) are optimal molecular diagnostic modalities in circumstances such as public health emergencies, rare disease diagnosis, limited budget, or where existing commercial alternatives are unavailable, limited in supply, or withdrawn, either temporarily or permanently. These tests reduce access barriers and enhance equitable clinical practice and healthcare delivery. Despite recommendations for the development of nucleic acid amplification tests, procedural details are often insufficient, inconsistent, and arbitrary. This protocol elucidates the methodology used in the development of a fully automated real-time polymerase chain reaction (qPCR)-based test, using the Panther Fusion® Open AccessTM functionality, for the detection of Streptococcus agalactiae in pregnant women, using selectively enriched rectovaginal swabs. In addition, guidelines are provided for oligonucleotide design (primers and TaqMan probes), in silico and in vitro evaluation of design effectiveness, optimization of the physicochemical conditions of the amplification reaction, and result analysis based on experimental designs and acceptance criteria. Furthermore, recommendations are provided for the analytical and clinical validation of the intended use. Our approach is cost-effective, particularly during the design and optimization phases. We primarily used open-source bioinformatics software and tools for in silico evaluations for the test design. Subsequently, the process was manually optimized using a CFX96 Dx analyzer, whose technical specifications and performance are homologous to that of the final platform (Panther Fusion®). Unlike Panther Fusion®, the CFX96 Dx does not require excess volumes of reagents, samples, and evaluation materials (dead volume) to accommodate potential robotic handling-associated imprecisions. The utilization of the CFX96 Dx analyzer represents a strategic approach to enhancing the efficiency of resources and the optimization of time during LDT optimization.
Key features
• Efficient and robust use of bioinformatics tools for designing primers and TaqMan probes for qPCR-based detection of Streptococcus agalactiae.
• Optimization of physicochemical conditions and evaluation of the design effectiveness of a qPCR-based laboratory-developed test (LDT).
• Analytical and clinical validation of the qPCR-based LDT developed on an open-access functionality (Open AccessTM) of an automated in vitro diagnostic platform, Panther Fusion® (Hologic).
Keywords: Laboratory-developed test (实验室自建检测)Graphical overview
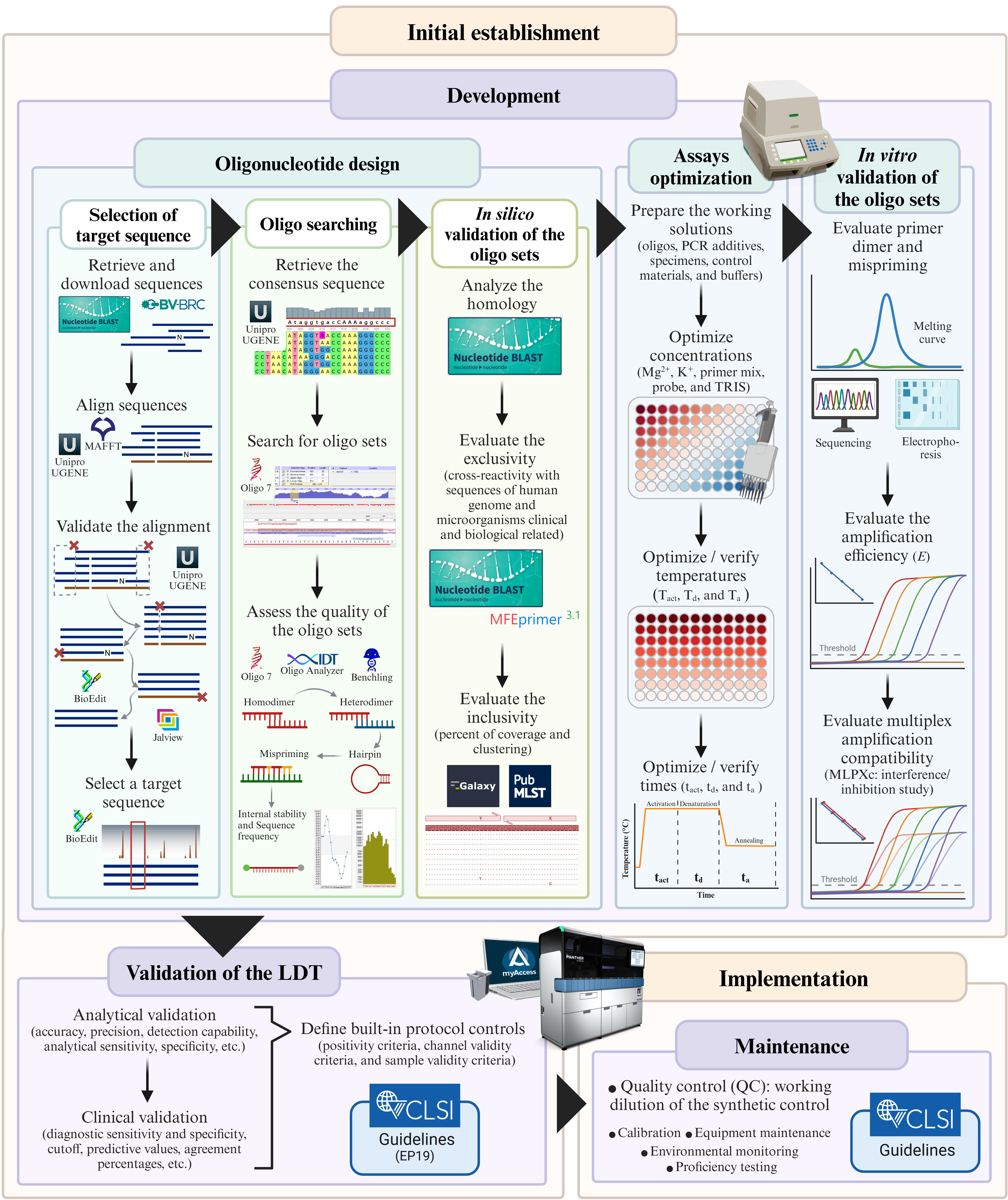
Protocol to develop a qPCR-based laboratory-developed test (LDT) for detecting Streptococcus agalactiae using the Panther Fusion® Open AccessTM system. Created in BioRender. Caballero Méndez, A. (2025) https://BioRender.com/n51j599.
Background
The Clinical and Laboratory Standards Institute (CLSI) defines a laboratory-developed test (LDT) as any test that has been designed, manufactured, and used within a single institution [1]. The use of LDTs is justified in scenarios where diagnostic tests are needed for rare and emerging infections, when commercial alternatives are unavailable, limited in supply, or withdrawn, or when cost-reduction strategies are needed in resource-limited settings. Validation of LDTs on automated in vitro diagnostic (IVD) platforms can potentially enhance laboratory responsiveness and efficiency, reduce access barriers, and improve equitable clinical practices and healthcare delivery. The Panther Fusion® system (Hologic, CA, USA) is an example of this potential. Its Open AccessTM functionality, fully automated sample-to-result processing, and continuous and random access capabilities enable high throughput and facilitate the safe implementation of qPCR-based LDTs [2]. In addition, bidirectional connectivity with laboratory information systems eliminates potential transcription errors [3].
However, LDT optimization on the Panther Fusion® Open AccessTM functionality is disadvantaged primarily by its high cost, largely attributable to the low yield of primer and probe reconstitution solutions (PPRs) and the substantial volumes of required samples, evaluation materials, or nucleic acid solutions. During optimization, multiple combinations of amplification mixture component concentrations must be prepared as PPRs and distributed within the system’s Open Access Packs. To compensate for imprecisions inherent to robotic handling of liquids, each PPR requires a minimum of 33% and a maximum of 70% more reactions than are strictly necessary. Hence, a convenient strategy for optimizing resources during this phase is the use of third-party qPCR analyzers that are similar to the Panther Fusion® in terms of detection channels and performance. Although undocumented for the Panther Fusion® Open AccessTM functionality, this strategy has been recommended for the open channels of other closed platforms, including the cobas® 5800/6800/8800 system [4,5].
Antepartum maternal rectovaginal colonization by Streptococcus agalactiae (Group B Streptococcus, GBS) is the leading risk factor for neonatal early-onset invasive GBS disease, which occurs within the first 6 days of life [6,7]. Culture remains the gold standard for universal screening; however, its limitations in sensitivity and turnaround time [8–10] have driven the development of alternative assays, such as those based on nucleic acid amplification (NAAT), which offer superior performance [11,12]. Herein, a protocol for the development of a qPCR-based LDT on the Panther Fusion® Open AccessTM functionality for the automated detection of the surface immunogenic protein (sip) gene of S. agalactiae is outlined. Furthermore, instructions for optimizing the assay’s physicochemical conditions using the CFX96 Dx analyzer (Bio-Rad, CA, USA) are provided. The evaluation of the analytical and clinical performance of the LDT, described herein as a validator of the protocols, is based on the recommendations of the CLSI, United States.
Materials and reagents
Biological materials
1. Streptococcus agalactiae Lehmann and Neumann (ATCC, 12386)
2. LIM broth-enriched rectovaginal swabs (clinical samples from women at ≥36 0/7 weeks’ gestation)
Reagents
1. Tris-ethylenediaminetetraacetic acid (10 mM Tris and 0.1 mM EDTA), pH 8.0 (IDTE buffer, pH 8.0) (IDT, catalog number: 11–05-01-13)
2. Tris-ethylenediaminetetraacetic acid (10 mM Tris and 0.1 mM EDTA), pH 7.5 (IDTE buffer, pH 7.5) (IDT, catalog number: 11-05-01-15)
3. 1 M Tris buffer (TRIS) (Hologic, catalog number: PRD-04935)
4. 1 M magnesium chloride (MgCl2) (Hologic, catalog number: PRD-04926)
5. 1 M potassium chloride (KCl) (Hologic, catalog number: PRD-04927)
6. Nuclease-free water (IDT, catalog number: 11-05-01-14)
7. iTaq Universal SYBR® green supermix (Bio-Rad, catalog number: 1725120)
8. Panther Fusion® Open AccessTM RNA/DNA enzyme cartridge (Hologic, catalog number: PRD-04303)
9. Panther Fusion® extraction reagents-X (Hologic, catalog number: PRD-04477)
10. Panther Fusion® internal control-X (IC-X) (Hologic, catalog number: PRD-04476)
11. eSwab® (Copan, catalog number: 480CE)
12. LIM broth, 5 mL (Todd Hewitt broth, 30 g/L; yeast extract, 10 g/L; nalidixic acid, 15 mg/L; and colistin sulfate, 10 mg/L) (Hardy Diagnostics, catalog number: L57)
13. Specimen transport medium (STM) (Hologic, catalog number: PRD-04423)
14. Aptima Specimen Transfer kit (AST) (Hologic, catalog number: 301154C)
15. DNA IC primers (Hologic, catalog number: PRD-04306)
16. DNA IC probe (Quasar 705) (Hologic, catalog number: PRD-04308)
17. Panther Fusion® elution buffer (Hologic, catalog number: PRD-04334)
18. Panther Fusion® oil (Hologic, catalog number: PRD-04335)
19. Oil reagent (Hologic, catalog number: PRD-04304)
20. Aptima auto detect (Hologic, catalog number: 303013)
21. Aptima Assay Fluids kit (Hologic, catalog number: 303014)
22. GBS forward primer (HPLC-double-purified from IDT): AGT TTC TCT CAA TAC AAT TTY GGA AGG T
23. GBS reverse primer (HPLC-double-purified from IDT): GCT GGC GCA GAA GAA TAT GTC T
24. GBS TaqMan-probe (HPLC-double-purified from IDT): 6-FAM/CA ATC GTT G/ZEN/K TGC TGC TTC TGG TGT CA/IABkFQ
25. UltramerTM duplex control (standard desalting-purified from IDT): GTT TCT GTT GCA GAC CAA AAA GTT TCT CTC AAT ACA ATT TCG GAA GGT ATG ACA CCA GAA GCA GCA ACA ACG ATT GTT TCG CCA ATG AAG ACA TAT TCT TCT GCG CCA GCT TTG AAA TCA AAA GAA GTA TT
26. MagNA Pure 24 Total DNA Isolation kit (Roche Diagnostics, catalog number: 07658036001)
27. MagNA pure bacterial lysis buffer (Roche Diagnostics, catalog number: 06374921001)
28. Proteinase K, recombinant, PCR grade (Roche Diagnostics, catalog number: 03115828001)
29. ID broth, 4.5 mL (potassium chloride, 7.5 g/L; calcium chloride, 0.5 g/L; tricine glycine, 0.895 g/L; and polysorbate 80, 0.025%) (saline solution) (Beckton, Dickinson and Co., catalog number: 246001)
30. BBLTM infusion agar (blood agar) (Beckton, Dickinson and Co., catalog number: 211037)
Solutions
1. Primer and probe reconstitution solution (PPR) for optimizing MgCl2 and KCl concentrations (see Recipes)
2. PPR for optimizing the concentration of the primer mix and probe for the LDT-GBS assay (see Recipes)
3. PPR for optimizing TRIS concentration (see Recipes)
4. PPR for optimizing Ta (see Recipes)
5. PPR for optimizing the primer mix and probe concentrations for an IC-X detection assay (see Recipes)
6. Optimized primer and probe reconstitution solution (oPPR) (see Recipes)
7. SYBR® green master mix (see Recipes)
Recipes
1. Primer and probe reconstitution solution (PPR) for optimizing MgCl2 and KCl concentrations
| Reagent | Working concentration | Final concentration | Unitarian volume1 | Volume (four tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 10.5–14 µL | 42–56 µL |
| KCl | 1 M | Variable3 | 0–2.5 µL | 0–10 µL |
| MgCl2 | 100 mM | Variable4 | 0.25–1.25 µL | 1–5 µL |
| TRIS | 100 mM | 8 mM | 2 µL | 8 µL |
| GBS forward primer | 10 µM | 0.6 µM | 1.5 µL | 6 µL |
| GBS reverse primer | 10 µM | 0.6 µM | 1.5 µL | 6 µL |
| GBS probe | 10 µM | 0.3 µM | 0.75 µL | 3 µL |
| Total volume | n/a | n/a | 20 µL | 80 µL |
1An amplification volume of 25 µL is assumed [20 µL of master mix (MMX) + 5 µL of DNA template].
2A PPR volume of 80 µL yields four tests using the CFX96 Dx analyzer: two replicates and two additional excess reactions (“dead reactions”), one for PPR and one for MMX.
3MgCl2 concentrations of 1.0, 2.0, 3.0, 4.0, and 5.0 mM.
4KCl concentrations of 0, 25, 50, 75, and 100 mM.
The volumes presented have been calculated using the calculation template mentioned in File S5.
2. PPR for optimizing the concentration of the primer mix and probe for the LDT-GBS assay
| Reagent | Working concentration | Final concentration | Unitarian volume1 | Volume (four tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 10.8–13.8 µL | 43–55 µL |
| KCl | 1 M | 100 mM | 2.5 µL | 10 µL |
| MgCl2 | 100 mM | 3 mM | 0.75 µL | 3 µL |
| TRIS | 100 mM | 8 mM | 2 µL | 8 µL |
| GBS primer mix | 10 µM | Variable3 | 0.5–2.5 µL | 2–10 µL |
| GBS probe | 10 µM | Variable4 | 0.5–1.5 µL | 2–6 µL |
| Total volume | n/a | n/a | 20 µL | 80 µL |
1An amplification volume of 25 µL is assumed (20 µL of MMX + 5 µL of DNA template).
2A PPR volume of 80 µL yields four tests using the CFX96 Dx analyzer: two replicates and two dead reactions (one for PPR and one for MMX).
3Primer mix concentrations of 0.2, 0.4, 0.6, 0.8, and 1.0 µM (1:1 ratio).
4Probe concentrations of 0.2, 0.3, 0.4, 0.5, and 0.6 µM.
The volumes presented have been calculated using the calculation template mentioned in File S5.
3. PPR for optimizing TRIS concentration
| Reagent | Working concentration | Final concentration | Unitarian volume1 | Volume(four tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 10.25–11.5 µL | 41–46 µL |
| KCl | 1 M | 100 mM | 2.5 µL | 10 µL |
| MgCl2 | 100 mM | 3 mM | 0.75 µL | 3 µL |
| TRIS | 100 mM | Variable3 | 1.25–2.5 µL | 5–10 µL |
| GBS forward primer | 10 µM | 0.6 µM | 1.5 µL | 6 µL |
| GBS reverse primer | 10 µM | 0.6 µM | 1.5 µL | 6 µL |
| GBS probe | 10 µM | 0.4 µM | 1 µL | 4 µL |
| Total volume1 | n/a | n/a | 20 µL | 80 µL |
1An amplification volume of 25 µL is assumed (20 µL of MMX + 5 µL of DNA template).
2A PPR volume of 80 µL yields four tests using the CFX96 Dx analyzer: two replicates and two dead reactions (one for PPR and one for MMX).
3TRIS concentrations of 5.0, 8.0, and 10 mM.
The volumes presented have been calculated using the calculation template mentioned in File S6.
4. PPR for optimizing Ta
| Reagent | Stock concentration | Final concentration | Unitarian volume1 | Volume(27.5 tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 16.83 µL | 462.6 µL |
| KCl | 1 M | 100 mM | 2.5 µL | 68.8 µL |
| MgCl2 | 1 M | 3 mM | 0.075 µL | 2.1 µL |
| TRIS | 1 M | 8 mM | 0.2 µL | 5.5 µL |
| GBS forward primer | 100 µM | 0.6 µM | 0.15 µL | 4.1 µL |
| GBS reverse primer | 100 µM | 0.6 µM | 0.15 µL | 4.1 µL |
| GBS probe | 100 µM | 0.4 µM | 0.1 µL | 2.8 µL |
| Total volume | n/a | n/a | 20 µL | 550 µL |
1An amplification volume of 25 µL is assumed (20 µL of MMX + 5 µL of DNA template).
2A PPR volume of 550 µL yields 27.5 tests: 24 replicates and 3.5 dead reactions.
The volumes presented have been calculated using the calculation template mentioned in File S7.
5. PPR for optimizing the primer mix and probe concentrations for an IC-X detection assay
| Reagent | Working concentration | Final concentration | Unitarian volume1 | Volume(four tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 2.08–11.43 µL | 8.33–45.67 µL |
| KCl | 1 M | 100 mM | 2.5 µL | 10 µL |
| MgCl2 | 100 mM | 3 mM | 0.75 µL | 3 µL |
| TRIS | 100 mM | 8 mM | 2 µL | 8 µL |
| DNA IC primer mix | 3.75 µM3 | Variable4 | 1.33–6.67 µL | 5.33–26.67 µL |
| DNA IC probe | 2.5 µM3 | Variable5 | 2–6 µL | 8–24 µL |
| Total volume | n/a | n/a | 20 µL | 80 µL |
1An amplification volume of 25 µL is assumed (20 µL of MMX + 5 µL of DNA template).
2A PPR volume of 80 µL yields four tests using the CFX96 Dx analyzer: two replicates and two dead reactions (one for PPR and one for MMX).
31:10 dilution of the factory preparation.
4Primer mix concentrations of 0.2, 0.4, 0.6, 0.8, and 1.0 µM (1:1 ratio).
5Probe concentrations of 0.2, 0.3, 0.4, 0.5, and 0.6 µM.
The volumes presented have been calculated using the calculation template mentioned in File S5.
6. Optimized primer and probe reconstitution solution (oPPR)
| Reagent | Working concentration | Final concentration | Unitarian volume3 | Volume(24 tests)4 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 16.43 µL | 697.9 µL |
| KCl | 1 M | 100 mM | 2.5 µL | 106.3 µL |
| MgCl2 | 1 M | 3 mM | 0.075 µL | 3.2 µL |
| TRIS | 1 M | 8 mM | 0.2 µL | 8.5 µL |
| GBS forward primer | 100 µM | 0.6 µM | 0.15 µL | 6.4 µL |
| GBS reverse primer | 100 µM | 0.6 µM | 0.15 µL | 6.4 µL |
| GBS probe | 100 µM | 0.4 µM | 0.1 µL | 4.3 µL |
| DNA IC primers1 | 37.5 µM | 0.3 µM | 0.2 µL | 8.5 µL |
| DNA IC probe2 | 25 µM | 0.2 µM | 0.2 µL | 8.5 µL |
| Total volume | n/a | n/a | 20 µL | 850 µL |
1,2Analyte specific reagent (ASR): Primer mix and a TaqMan-probe, respectively, for detecting the Panther Fusion® internal control-X (IC-X).
3An amplification volume of 25 µL is assumed (20 µL of MMX + 5 µL of DNA template).
4A PPR volume of 850 µL yields 24 tests on the Panther Fusion® system (excluding the dead space volume) and requires 350 µL of oil reagent.
The volumes presented have been calculated using the calculation template mentioned in File S7.
7. SYBR® green master mix
| Reagent | Working concentration | Final concentration | Unitarian volume1 | Volume(24 tests)2 |
|---|---|---|---|---|
| Nuclease-free water | n/a | n/a | 4.7 µL | 117.5 µL |
| GBS forward primer | 100 µM | 0.6 µM | 0.15 µL | 3.75 µL |
| GBS reverse primer | 100 µM | 0.6 µM | 0.15 µL | 3.75 µL |
| SYBR® green supermix3 | 2× | 1× | 10 µL | 250 µL |
| Total volume | n/a | n/a | 15 µL | 375 µL |
1An amplification volume of 20 µL is assumed (15 µL of MMX + 5 µL of DNA template).
2A PPR volume of 375 µL yields 24 tests on the CFX96 Dx analyzer: eight levels × three replicates and one dead reaction.
3From iTaq Universal SYBR® green supermix.
Laboratory supplies
1. Hard-shell PCR plates, 96 well, thin wall (Bio-Rad, catalog number: HSP9655)
2. MicroAmp® optical adhesive film (Applied Biosystems, catalog number: 4311971)
3. MicroAmp® thin-walled reaction tube with flat cap, 0.5 mL (Applied Biosystems, catalog number: N8010737)
4. SC micro tube PCR-PT, 1.5 mL (Sarstedt, catalog number: 72.692.405)
5. SC micro tube PCR-PT, 2.0 mL (Sarstedt, catalog number: 72.693.465)
6. Multi-tube units (MTU) (Hologic, catalog number: 104772-02)
7. Panther Fusion® tube trays (Hologic, catalog number: PRD-04000)
8. Tips, 1,000 µL filtered, conductive, liquid-sensing, and disposable (Tecan, catalog number: 10612513)
9. Specimen aliquot tubes (Hologic, catalog number: 503762)
10. (Optional) Aptima® penetrable caps (Hologic, catalog number: 105668)
11. Panther waste bag kit (Hologic, catalog number: 902731)
12. Panther waste bin cover (Hologic, catalog number: 504405)
13. Disposable filter tips, nuclease-free certified, 10, 20, 50, 100, 200, and 1,000 µL (any manufacturer)
14. Micrewtubes with low adhesion surface, 2 mL, self-standing (PPR tubes) (Simport, catalog number: T341-6TLST100)
15. Screw caps for microtubes with O-ring, neutral color (caps for PPR tubes) (Simport, catalog number: T340NOS100)
16. (Optional) ChillBlockTM tube racks for MCT (Simport Scientific Inc., catalog number: S700-14)
17. MagNA Pure 24 processing cartridge (Roche Diagnostics, catalog number: 07345577001)
18. MagNA Pure 24 processing tip park/piercing tool (Roche Diagnostics, catalog number: 07345585001) or MagNA Pure 24 piercing tool (Roche Diagnostics, catalog number: 07534205001)
19. MagNA Pure tip 1,000 µL (Roche Diagnostics, catalog number: 06241620001)
20. MagNA Pure tip waste tray (Roche Diagnostics, catalog number: 08185492001)
21. MagNA Pure tube 2.0 mL (Roche Diagnostics, catalog number: 07857551001)
22. MagNA Pure sealing foil (Roche Diagnostics, catalog number: 06241638001)
Equipment
1. CFX96 Dx system [Bio-Rad, catalog numbers: 1845097-IVD (CFX96 Dx ORM) and 1841000-IVD (C1000 Dx Thermal Cycler)]
2. Panther Fusion® Open Access® system (Hologic, catalog number: PRD-04172)
3. MagNA Pure 24 instrument (Roche Diagnostics, catalog number: 07290519001)
4. Digital vortex mixer (Thermo Scientific, catalog number: 88882009)
5. SorvallTM ST 8 small benchtop centrifuge (Thermo Scientific, catalog number: 75007200)
6. Buckets for Thermo ScientificTM M10 microplate swinging bucket rotor (Thermo Scientific, catalog number: 75005723)
7. MicroClick 24 × 2 fixed angle microtube rotor (Thermo Scientific, catalog number: 75005715)
8. MALDI Biotyper®-BD Sirius IVD system (Bruker, catalog number: 1875321)
9. BD PhoenixTM AP instrument (Beckton, Dickinson and Co., catalog number: 448010)
Software and datasets
1. Unipro UGENE v48.0 or newer versions (Unipro, Novosibirsk, Russia, 09/08/2023) [13]
2. Jalview v2.11.2.7 or newer versions (University of Dundee, Scotland, UK, 06/30/2023) [14]
3. BioEdit v7.7.1 or newer versions (Tom Hall, 05/10/2021) [15]
4. Oligo v7.60 (Molecular Biology Insights, Inc., CO, USA, 12/2018) [16]
5. BDAL v12.0 library (11 897 MSP, rev. 2023)
6. MBT Compass (RUO/GP) v4.1.100 (Bruker, MA, USA)
7. myAccessTM v2.1.2.1 or newer versions (Hologic, CA, USA, 06/08/2022)
8. Analyse-it for Microsoft Excel, Ultimate Edition v6.15.4 or newer versions (Analyse-it Software Ltd., Leeds, UK, 04/18/2023)
9. Microsoft Excel 2016 or newer versions (Microsoft Corporation)
10. Bacterial and Viral Bioinformatics Resource Center (BV-BRC, University of Chicago, USA) (https://www.bv-brc.org) (access date, 02/10/2022)
11. GenBank database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genbank/) (access date, 02/12/2022)
12. Basic Local Alignment Search Tool (BLAST) of the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (access date, 04/20/2022)
13. Entropy Calculator tool (The National Institute of Public Health, Czech Republic) (https://entropy.szu.cz/EntropyCalcWeb/entropy) (access date, 02/25/2022) [17]
14. Multiple Alignment using the Fast Fourier Transform (MAFFT, Osaka University, Japan) (http://mafft.cbrc.jp/alignment/server/) (access date, 03/03/2022) [18]
15. BenchlingTM (Benchling, CA, USA) (https://benchling.com/) (access date, 03/08/2022)
16. OligoAnalyzerTM tool (IDT, IA, USA) (https://www.idtdna.com/calc/analyzer) (access date, 03/08/2022)
17. MFEprimer, v3.1 (iGeneTech Bioscience, Beijing, China) (https://mfeprimer3.igenetech.com/) (access date, 05/08/2022) [19,20]
18. Public Databases for Molecular Typing and Microbial Genome Diversity (PubMLST, University of Oxford, UK) (https://www.pubmlst.org/) (access date, 05/12/2022) [21]
19. Bacterial Isolate Genome Sequence Database online software (BIGSdb, University of Oxford, UK) (access date, 05/12/2022) [22]
20. Polymerase Chain Reaction Evaluation Through Large-Scale Mining of Genomic Data (SCREENED) v1.0 (Sciensano Galaxy External) (https://galaxy.sciensano.be/) (access date, 05/13/2022) [23]
21. Resuspension Calculator (IDT, IA, USA) (https://www.idtdna.com/Calc/resuspension/) (access date, 06/05/2022)
22. Dilution Calculator (IDT, IA, USA) (https://www.idtdna.com/Calc/Dilution/) (access date, 06/05/2022)
23. WinEpi v2.0 (University of Zaragoza, Spain) (http://www.winepi.net/winepi2/f102.php) (access date, 04/05/2022) [24]
Procedure
文章信息
稿件历史记录
提交日期: Dec 31, 2024
接收日期: Feb 25, 2025
在线发布日期: Mar 11, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Caballero Méndez, A., Reynoso de La Rosa, R. A., Abreu Bencosme, M. E., Sosa Ortiz, M. N., Pichardo Beltré, E., de La Cruz García, D. M., Piñero Santana, N. J. and Bacalhau de León, J. C. (2025). Screening for Streptococcus agalactiae: Development of an Automated qPCR-Based Laboratory-Developed Test Using Panther Fusion® Open AccessTM. Bio-protocol 15(7): e5255. DOI: 10.21769/BioProtoc.5255.
分类
生物信息学与计算生物学
分子生物学 > DNA > DNA 检测
微生物学 > 病原体检测 > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link















