- EN - English
- CN - 中文
A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma
肺腺癌中激光显微切割与tRF/tiRNA测序操作方法
(*Contributed equally to this work, §Technical contact: wangzi@stu.njmu.edu.cn, **Lead contact) 发布: 2025年04月05日第15卷第7期 DOI: 10.21769/BioProtoc.5253 浏览次数: 3004
评审: Ritu GuptaSalah BoudjadiNavnita DuttaHeng Sun
Abstract
Laser-assisted microdissection (LAM) coupled with next-generation sequencing technologies offers a powerful approach to dissecting the complex cellular heterogeneity within lung adenocarcinoma (LUAD) tumors. This protocol outlines the method for isolating specific high-risk LUAD tissues containing micropapillary/solid (MIP/SOL) patterns, which is linked to poor prognosis. We detail the process of LAM, which involves tissue fixation, microtome sectioning, and the precise dissection and collection of cells of interest under microscopic guidance. The isolated cells are then subjected to RNA extraction, library preparation, and sequencing to profile transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs), which are emerging as key regulators in cancer. This protocol enables researchers to obtain high-quality transcriptomic data from specific LUAD cell populations, aiming to uncover tRF-Val-CAC-024 and tiRNA-Gly-CCC as potential biomarkers for early diagnosis and therapeutic targets for LUAD treatment.
Key features
• Laser-assisted microdissection (LAM): Utilizes laser precision to isolate high-risk MIP/SOL patterns of LUAD tissues.
• tRF & tiRNA sequencing: Profiles transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs), reported to play a key role in cancer development.
Keywords: Laser microdissection (激光显微切割)Graphical overview
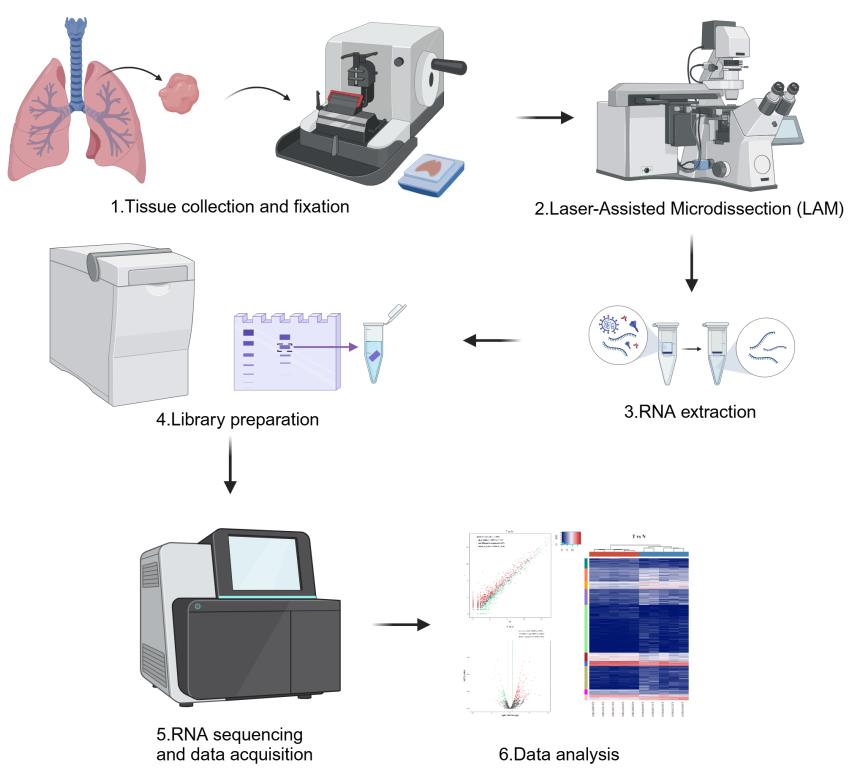
Background
Lung adenocarcinoma (LUAD), a subtype of non-small cell lung cancer, is characterized by its molecular heterogeneity and complex metastatic behavior. Understanding the molecular underpinnings of LUAD is crucial for advancing our knowledge of cancer biology and for the development of targeted therapies. Traditional bulk sequencing approaches provide a comprehensive view of the transcriptome but often mask the cellular heterogeneity within tumors [1]. There is a need for techniques that can isolate and characterize the transcriptomic profiles of specific cell populations within LUAD tumors. Laser-assisted microdissection (LAM) is a microscopy-based technique that allows for the precise isolation of specific cell populations directly from their tissue context. This method has been increasingly used in oncology research to study the molecular signatures of distinct cell types within tumors. LAM offers several advantages over other methods, such as fluorescent activated cell sorting (FACS), as it does not require the generation of marker lines or prior knowledge of a large number of marker genes. Instead, LAM relies solely on the morphological characteristics of the tissues to be collected, making it a versatile tool for studying various cancer types, including LUAD [2]. The combination of LAM with next-generation sequencing has emerged as a powerful approach to dissecting the complex cellular heterogeneity within LUAD tumors. In particular, the study of transfer RNA–derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs) has been gaining momentum, as these small RNA species are implicated in various biological processes, including gene regulation, protein translation, and cellular metabolism, which are often dysregulated in cancer [3].
In this protocol, we describe the method for LAM of LUAD tissues to isolate high-risk components containing micropapillary/solid (MIP/SOL) patterns, which are known to be associated with poor prognosis. We further detail the process of tRF and tiRNA sequencing to characterize the molecular signatures of these high-risk LUAD cell populations. This approach not only enhances our understanding of LUAD biology but also paves the way for the identification of novel therapeutic targets and biomarkers [4].
By following this protocol, researchers can effectively isolate and analyze the transcriptomic profiles of specific LUAD cell populations, contributing to the advancement of personalized medicine. The protocol is designed to be adaptable for various research applications, including the study of other cancer types and complex tissue samples.
Materials and reagents
Biological materials
1. Lung adenocarcinoma (LUAD) tissues and adjacent normal tissues collected during thoracoscopic surgery
Reagents
1. 10% neutral buffered formalin (NBF) (Sigma-Aldrich, catalog number: HT501128) for tissue fixation
2. Xylene (Thermo Fisher Scientific, catalog number: X3S-4)
3. Absolute ethyl alcohol (ethanol) (Sigma-Aldrich, catalog number: E7023)
4. HistoClear II (National Diagnostics, catalog number: HS-202) for tissue clearing and processing
5. Paraplast Plus® (paraffin) (Leica Biosystems, catalog number: 39601006) for tissue embedding
6. Trypsin-EDTA (Thermo Fisher Scientific, catalog number: 25200056) for cell disassociation (if single-cell sequencing is required)
7. PureLink RNA Micro kit (Thermo Fisher Scientific, catalog number: 12183016) for total RNA extraction
8. NextSeq 500/550 v2 kit (Illumina, catalog number: FC-404-2005) for tRF and tiRNA sequencing
9. Small RNA adapters (Illumina, catalog number: 15004197)
Laboratory supplies
1. Sterile Falcon tubes (for sample storage) (Corning, catalog number: 352096)
2. Beakers (for solution mixing) (Pyrex, catalog number: 1000-100)
3. Dissecting forceps [for manual tissue dissection (if not using LAM)] (Fine Science Tools, catalog number: 11050-10)
4. Steel inclusion molds (for tissue embedding) (Leica Biosystems, catalog number: 14046435645)
5. Embedding cassettes without the lid (for tissue embedding) (Thermo Fisher Scientific, catalog number: 15182731)
6. Microscope slides (for tissue sectioning) (Thermo Fisher Scientific, catalog number: 12-550-15)
7. Low-profile disposable blades (for tissue sectioning) (Feather Safety Razor Co., catalog number: 0915)
8. Slide rack and glass staining dish with lid (for tissue staining and deparaffinization) (Thermo Fisher Scientific, catalog number: S90010)
9. TubeCollector 2×200 CM II (for collecting laser micro-dissected tissues) (Zeiss, catalog number: 415190-9081-000)
10. AdhesiveCap 200 clear (for sealing microcentrifuge tubes containing micro-dissected tissues) (Zeiss, catalog number: 415190-9191-000)
11. MembraneSlide 1.0 OPEN (for tissue section adhesion and slide preparation) (Zeiss, catalog number: 415190-9051-000)
12. RNase-free CapSure Macro caps (Zeiss, catalog number: 415190-9191-000)
13. RNase-free wipes (Thermo Fisher Scientific, catalog number: 21-404-303)
Equipment
1. Vacuum pump (Thermo Fisher Scientific, model: R4100649 or Edwards, model: E2M28)
2. Vacuum desiccator (Bel-Art, catalog number: 42022000 or Cole-Parmer, catalog number: EW-06100-20)
3. Mini rotator (Benchmark Scientific, catalog number: H5500 or Thermo Fisher Scientific, model: 88881001)
4. Convection incubator (Thermo Scientific, model: Heratherm IMC18 or Memmert, model: UF110)
5. Paraffin dispenser (Leica Biosystems, model: HistoCore Arcadia H)
6. Microtome (Leica Biosystems, model: RM2235 or Thermo Fisher Scientific, model: HM325)
7. Slide warmer (Leica Biosystems, model: HI1220 or Thermo Fisher Scientific, catalog number: 22-050-263)
8. PALM MicroBeam IV microscope with motorized RoboMover and CapMover (Zeiss, model: PALM MicroBeam IV)
9. NanoDrop (optional) (Thermo Fisher Scientific, model: NanoDrop 2000, NanoDrop One)
10. Covaris S2 ultrasonicator (optional) (Covaris, model: S2)
11. Qubit 4 fluorometer (optional) (Thermo Fisher Scientific, catalog number: Q33238)
12. Agilent Bioanalyzer (Agilent Technologies, model: 2100 Bioanalyzer)
13. Automated gel cutter (Blue-Ray Biotech, catalog number: BLU0001)
Software and datasets
1. PALM Robo software (Zeiss, v4) for operating the LCM system
2. Optional: R studio for RNA-seq data analysis
3. Bioinformatics tools for RNA-seq data processing and analysis, including FastQC, cutadapt, bowtie, and edgeR
4. tRNAscan-SE (v2.0.11, http://lowelab.ucsc.edu/tRNAscan-SE/)
Procedure
文章信息
稿件历史记录
提交日期: Nov 23, 2024
接收日期: Feb 17, 2025
在线发布日期: Mar 10, 2025
出版日期: Apr 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Wang, Z., Wang, Q., Xu, L., Mao, Q. and Jiang, F. (2025). A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma. Bio-protocol 15(7): e5253. DOI: 10.21769/BioProtoc.5253.
分类
癌症生物学 > 癌症生物化学 > 癌代谢
系统生物学 > 转录组学 > RNA测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













