- EN - English
- CN - 中文
Improved Extraction Methods to Isolate High Molecular Weight DNA From Magnaporthaceae and Other Grass Root Fungi for Long-Read Whole Genome Sequencing
优化高分子量 DNA 提取方法以用于 Magnaporthaceae 及其他禾本科根部真菌的长读长全基因组测序
发布: 2025年03月20日第15卷第6期 DOI: 10.21769/BioProtoc.5245 浏览次数: 3230
评审: Xiaofei LiangAnonymous reviewer(s)

相关实验方案

用于跨谱应急和现场使用的快速、可持续的热渗透 DNA 提取方案
Stavroula Goudoudaki [...] Yiannis Manoussopoulos
2023年09月05日 1715 阅读
Abstract
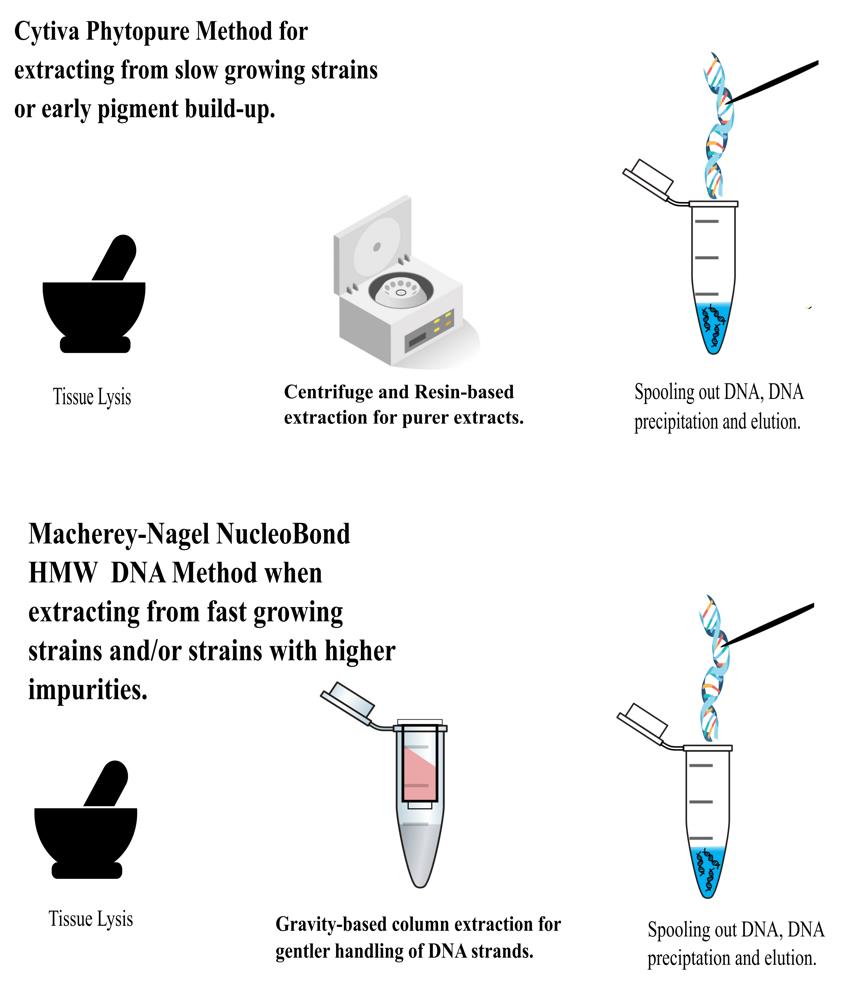
This manuscript details two modified protocols for the isolation of long-stranded or high molecular weight (HMW) DNA from Magnaporthaceae (Ascomycota) fungal mycelium intended for whole genome sequencing. The Cytiva Nucleon PhytoPure and the Macherey-Nagel NucleoBond HMW DNA kits were selected because the former requires lower amounts of starting material and the latter utilizes gentler methods to maximize DNA length, albeit at a higher requirement for input material. The Cytiva Nucleon PhytoPure kit successfully recovered HMW DNA for half of our fungal species by increasing the amount of RNase A treatment and adding in a proteinase K treatment. To reduce the impact of pigmentation development, which occurs toward later stages of culturing, extractions were run in quadruplicate to increase overall DNA concentration. We also adapted the Macherey-Nagel NucleoBond HMW DNA kit for high-quality HMW DNA by grinding the sample to a fine powder, overnight lysis, and splitting the sample before washing the precipitated DNA. For both kits, precipitated DNA was spooled out pre-washing, ensuring a higher percentage of high-integrity long strands. The Macherey-Nagel protocol offers advantages over the first through the utilization of gravity columns that provide gentler treatment, yielding >50% of high-purity DNA strands exceeding 40 kbp. The limitation of this method is the requirement for a large quantity of starting material (1 g). By triaging samples based on the rate of growth relative to the accumulation of secondary metabolites, our methodologies hold promise for yielding reliable and high-quality HMW DNA from a variety of fungal samples, improving sequencing outcomes.
Key features
• Modified protocols for the extraction of high molecular weight fungal DNA suitable for long-read whole genome sequencing.
• Approximately 4 h to complete four samples in parallel (excluding lysis time).
• Optimized for the mycelium of Magnaporthaceae fungi, harvested before melanin (or secondary metabolite) buildup, but readily adaptable for other ascomycetes.
Keywords: Fungi (真菌)Graphical overview
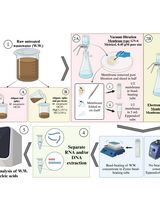
Extraction process
Background
High-quality fungal genome assemblies play a crucial role in understanding the evolution of virulence and identifying genetic factors associated with pathogenic and non-pathogenic lifestyles [1–3]. Short read data tends to result in genome assemblies that are more fragmented because reads are unable to span structural variants and repetitive regions [4]. Fungi may be particularly impacted by structural genomic variation and giant mobile elements, which may be underestimated by current assemblies [5–6]. As such, demand has emerged for the generation of long-read data better able to span repetitive regions of the genome to improve assembly contiguity and the incorporation of such repetitive content. Producing long-read sequencing data is dependent on high molecular weight (HMW) DNA. In fungi, extracting the high concentrations of DNA needed whilst also maintaining strand length and integrity is challenging, perhaps due to their cell walls, production of secondary metabolites, or the often small amounts of starting material [7]. Several kits and methods were explored before settling on modified methods using the Cytiva Nucleon PhytoPure and the Macherey-Nagel HMW DNA kits (see Table S1). Our aim was to extract HMW DNA for PacBio sequencing in a volume higher than 13 μg. Harvesting the mycelium before pigmentation caused by the buildup of melanin and other secondary metabolites improves DNA quality after extraction using both modified methods. We have developed and optimized HMW DNA extraction methodologies for Magnaporthaceae species and other grass root fungal species.
Part I
Cytiva Nucleon PhytoPure kit modified protocol
Materials and reagents
Reagents
1. Cytiva Nucleon PhytoPure, pack of 50 × 1.0 g (Cytiva, catalog number: RPN8511); this kit contains Reagent 1, Reagent 2, and Phytopure resin
2. Propanol-2-OL (Merck Life Sciences, catalog number: 33539-2.5LM, CAS 67-63-0)
3. Chloroform, analytical grade (Merck Life Sciences, catalog number: 32211-2.5LM, CAS 67-66-3)
4. Ethanol, denatured (VWR, catalog number: 01000940, CAS 64-17-5)
5. Invitrogen PureLink RNase A, 20 mg/mL (Fisher Scientific, catalog number: 10618703)
6. Proteinase K, 30 mg/mL (Thermo Scientific, catalog number: 11501515)
7. Low-TE elution buffer, 1× solution, pH 8.0 (Fisher Scientific, catalog number: 10647633)
8. Potato dextrose broth (Formedium, catalog number: PDB0102)
10. Qubit high sensitivity assay (Invitrogen, catalog number: Q23854)
11. Qubit broad range assay (Invitrogen, catalog number: Q32853)
Solutions
1. 70% ethanol solution (EtOH) (see Recipes)
Recipes
1. 70% EtOH
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol, 99.9% pure | n/a | 30 mL |
| Sterilized purified water | n/a | 20 mL |
| Total (optional) | n/a | 50 mL |
Note: This solution should be made up fresh on the day of the extraction.
Laboratory supplies
1. 2 mL DNA LoBind Eppendorf tubes (Fisher Scientific, catalog number: 10031282)
2. Filter p1000 tips (Elkay Laboratory Products, catalog number: AER-2REF-S96)
3. Filter p200 tips (Elkay Laboratory Products, catalog number: AER-REF-S96)
4. Paper tissue medical wipes (Bunzle Cleaning and Hygiene Supplies, catalog number: 066080)
5. Disposable PES bottle top filter 0.2 μm, 500 mL capacity (Fisher Brand, catalog number: 15983307)
6. 0.22 μm bottle top filter (Fisherbrand disposable PES) (Fisher Scientific, catalog number: 15993307)
Equipment
1. Thermomixer-mixer HC (Starlab Smart Instruments, model: S8012-0000)
2. 100–1,000 mL ErgoOne pipette (Starlabs, model: S7100-1000)
3. 20–200 mL ErgoOne pipette (Starlabs, model: S7100-2200)
4. Centrifuge (Eppendorf, model: 54520000060 mini spin plus)
5. 250 mL glazed mortar and pestle (Haldenwanger, model: 55/3/glazed)
6. Freezer -20 °C and freezer -80 °C
7. Vortex mixer (Stuart Science Equipment, model: SA8)
8. Vacuum pump (WooSung, model: W2v20)
9. Growth cabinet (Binder, model: BD400)
10. Orbital shaker (SciQuip, model: SP2250-07)
11. NanoDrop One Microvolume UV-Vis spectrophotometer (Thermo Scientific, model: NC-ONEC-W)
12. Qubit fluorometer (Invitrogen, model: Qubit 3.0, catalog number: Q32854)
13. Autoclave (Astell, model: ASB30019293)
14. Orbital Shaker (SciQuip model: shaker 07)
15. Growth cabinet (Binder, model: BD400)
16. Vacuum pump (JungWoo, model: Woo Sung Automa 20)
17. 500 mL borosilicate glass Erlenmeyer flasks (Fisher Scientific, catalog number: 15429113)
18. Femto Pulse system (Agilent, catalog number: M5330AA)
Procedure
文章信息
稿件历史记录
提交日期: Oct 18, 2024
接收日期: Jan 25, 2025
在线发布日期: Mar 10, 2025
出版日期: Mar 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Grey, M. J., Freeman, J., Rudd, J., Irish, N., Canning, G., Chancellor, T., Palma-Guerrero, J., Hill, R., Hall, N., Hammond-Kosack, K. E. and McMullan, M. (2025). Improved Extraction Methods to Isolate High Molecular Weight DNA From Magnaporthaceae and Other Grass Root Fungi for Long-Read Whole Genome Sequencing. Bio-protocol 15(6): e5245. DOI: 10.21769/BioProtoc.5245.
分类
微生物学 > 微生物遗传学 > 全基因组测序
分子生物学 > DNA > DNA 提取
系统生物学 > 基因组学 > 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link