- EN - English
- CN - 中文
Visualization of mRNA Translation Within Germ Granule Biphasic Organization in Drosophila Early Embryo
果蝇早期胚胎生殖颗粒双相结构中 mRNA 翻译的可视化
发布: 2025年03月20日第15卷第6期 DOI: 10.21769/BioProtoc.5242 浏览次数: 3589
评审: Marion HoggTing MiaoKyle Stewart Skalenko
Abstract
Super-resolution imaging of RNA–protein (RNP) condensates has shown that most are composed of different immiscible phases reflected by a heterogenous distribution of their main components. Linking RNA–protein condensate’s inner organization with their different functions in mRNA regulation remains a challenge, particularly in multicellular organisms. Drosophila germ granules are a model of RNA–protein condensates known for their role in mRNA storage and localized protein production in the early embryo. Present at the posterior pole of the embryo within a specialized cytoplasm called germplasm, they are composed of maternal mRNAs as well as four main proteins that play a key role in germ granule formation, maintenance, and function. Germ granules are necessary and sufficient to drive germ cell formation through translational regulation of maternal mRNAs such as nanos. Due to their localization at the posterior tip of the ovoid embryo and small size, the classical imaging setup does not provide enough resolution to reach their inner organization. Here, we present a specific mounting design that reduces the distance between the germ granule and the objectives. This method provides optimal resolution for the imaging of germ granules by super-resolution microscopy, allowing us to demonstrate their biphasic organization characterized by the enrichment of the four main proteins in the outermost part of the granule. Furthermore, combined with the direct visualization of nanos mRNA translation using the Suntag approach, this method enables the localization of translation events within the germ granule’s inner organization and thus reveals the spatial organization of its functions. This approach reveals how germ granules serve simultaneously as mRNA storage hubs and sites of translation activation during development. This work also highlights the importance of considering condensates’ inner organization when investigating their functions.
Key features
• Method for super-resolution imaging of germ granules in Drosophila early embryo.
• Analysis of RNP condensate functional organization.
• Simultaneous recording of RNP condensate function and organization.
Keywords: RNP condensates (RNP 凝聚体)Graphical overview
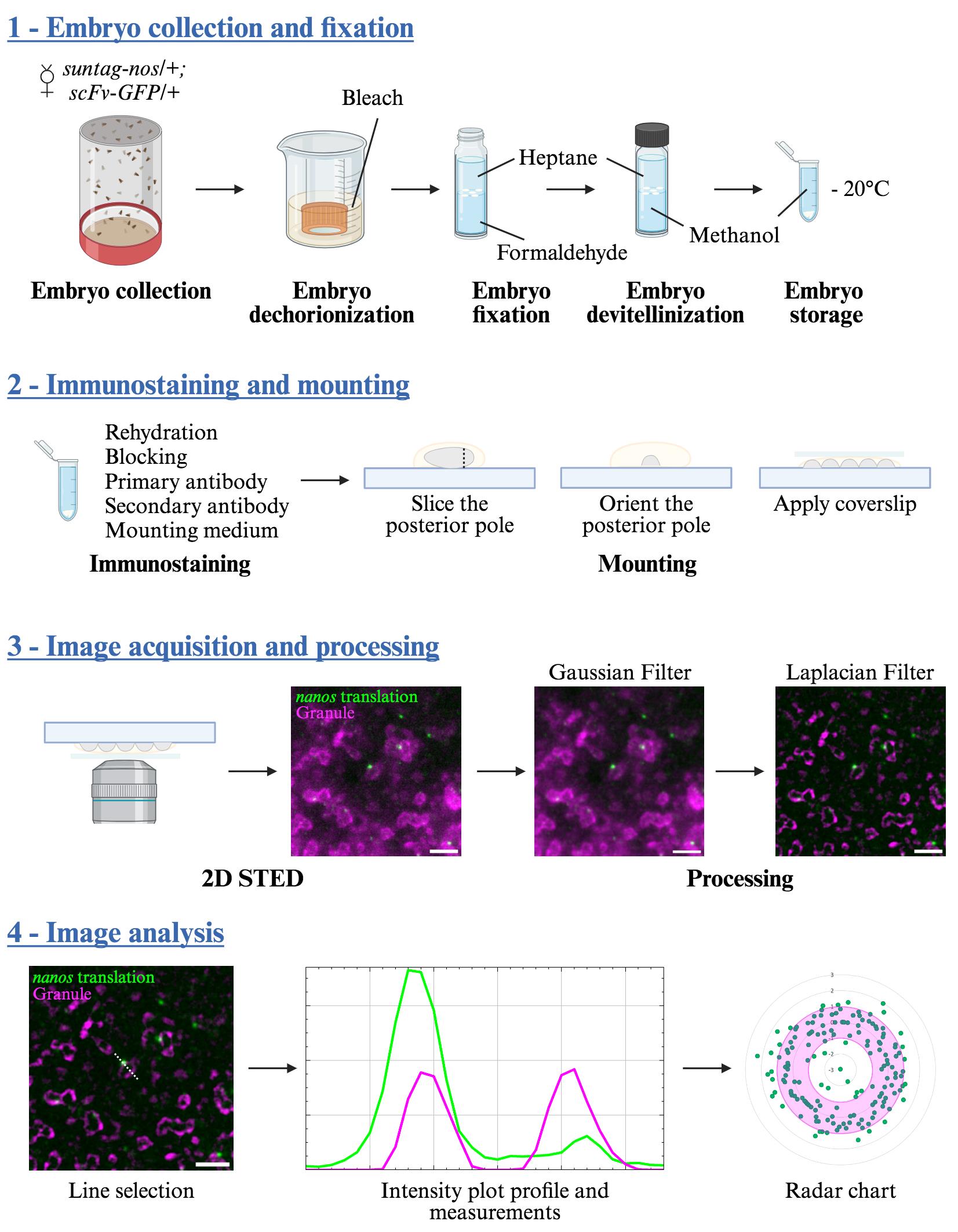
Visualization of mRNA translation within germ granule biphasic organization
Background
RNA–protein (RNP) condensates, also called RNA granules, exist in both the nucleus and cytoplasm and are linked to various aspects of RNA biology [1,2]. Most RNP condensates are composed of multiple immiscible phases, reflected by a heterogeneous distribution of RNA and/or RNA binding proteins [2,3]. Linking RNP condensate functions with their inner organization is a challenge that requires simultaneous recording of both their structure and functions.
Drosophila germ granules are a model of RNP condensates. Localized at the posterior pole of the embryo, they are necessary and sufficient to drive germ cell fate through maternal mRNA storage and their sequential translational activation [4,5]. Composed of four main proteins [Oskar (Osk), Vasa, Tudor, and Aubergine (Aub)] together with more than 200 maternal mRNAs such as nanos mRNA [5–7], they have long been suspected to be a site for localized mRNA translation [8]. We previously showed that Aub is involved in nanos mRNA translational activation through the recruitment of translation initiation factors at the edge of germ granules [9]. This led us to hypothesize that mRNA regulation is organized within the germ granule, with translation occurring at the surface while mRNA storage would be restricted to an internal region of the granule. We thus further explored germ granule inner structure and where mRNA translation and repression occurred in these condensates. However, germ granules have a relatively small size compared to most RNP condensates, ranging from 200 to 500 nm [8], thus rendering the use of super-resolution microscopy mandatory to reveal their organization. Another problem lies in the localization of germ granules at the posterior tip of the ovoid embryo. This implies imaging far from the microscope objective when the embryo lies on the slide, resulting in a decreased resolution not compatible with super-resolution microscopy. To render super-resolution imaging compatible with Drosophila early embryo, we set up a mounting design whereby the posterior pole of the embryo was sliced and placed with germ granules facing the coverslip. This reduced the distance between germ granules and the objective as well as sample-induced light scattering and absorption (the result of imaging deep in a tissue), thus substantially improving resolution. By using this method with stimulated emission depletion (STED) microscopy, whereby a depletion laser reversibly silences fluorophores around the center of the excitation focus, we were able to reach a resolution of 60 nm in our samples and thus reveal germ granule biphasic organization with enrichment of their main proteins in the outermost phase [10]. This mounting method is compatible with other imaging techniques such as AiryScan and OMX [10]; however, STED microscopy provides the best resolution. To record translation at germ granules, we implemented the Suntag technique [11–13] to visualize nanos mRNA translation. Briefly, binding a single-chain antibody fused to GFP (scFv-GFP) to repeats of Suntag epitopes localized downstream of the nanos start codon allows the detection of nascent peptides by creating bright GFP spots above the background. Combined with our method of super-resolution imaging, we were able to precisely localize translation events within the germ granule’s biphasic organization, showing that translation occurs in the outer phase (or shell) and at the surface of the granule and revealing that translational regulation at germ granule is compartmentalized.
Materials and reagents
Biological materials
1. Drosophila melanogaster stock scFv-GFP (w1118;; nos-scFv-GFP) [14]. The scFv-GFP flies contain a transgene encoding the single-chain antibody that recognizes the Suntag epitope fused to GFP and under the control of the germline-specific nanos promoter
2. Drosophila melanogaster stock suntag-nos (w1118; nos-suntagX12-nos/Cyo) generated in our lab [10]. The suntag-nos flies contain a transgene with a nanos genomic region containing 12 Suntag repeats localized after the start codon
3. Drosophila melanogaster stock w1118, referred to as wild type
Reagents
1. Commercial bleach 9.6% 250 mL (local purchase); store in the dark
2. Yeast (local purchase)
3. Formaldehyde 36%, AnalaR NORMAPUR (VWR, catalog number: 20909.290)
4. n-Heptane, AnalaR NORMAPUR (VWR, catalog number: 24551.290)
5. Methanol, AnalaR NORMAPUR (VWR, catalog number: 20847.295)
6. Dulbecco's phosphate-buffered saline (PBS) 10× (Sigma, catalog number: D1408)
7. Tween 20 (Sigma, catalog number: P1379)
8. Bovine serum albumin (BSA) (Sigma, catalog number: A2934)
9. Rabbit anti-Osk antibody (generated in our lab [10]); store at -20 °C in small aliquots to avoid multiple thaw cycles
10. Goat anti-rabbit IgG Abberior Star580 (Abberior, catalog number: ST580-1002-500UG); store at -20 °C in small aliquots to avoid multiple thaw cycles
11. FluoTag®-X4 anti-GFP Abberior StarRed (NanoTag Biotechnologies, catalog number: N0304-AbRED-L); store at -20 °C in small aliquots to avoid multiple thaw cycles
12. Abberior MOUNT, liquid antifade (Abberior, catalog number: MM-2009); store at -20 °C in small aliquots to avoid multiple thaw cycles
13. Neutral Red dye (Sigma, catalog number: N7005)
14. Fly culture medium in tubes or bottles [for 1 L: 8.75 g of agar, 75 g of yeast, 75 g of corn flour, 49.5 mL of Moldex; provided by a local platform (DROSO, Biocampus, Montpellier)]
Solutions
1. 2.6% bleach (see Recipes)
2. 10% BSA (see Recipes)
3. PBST (see Recipes)
4. Methanol dilutions (see Recipes)
Recipes
1. 2.6% bleach
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Bleach 9.6% | 2.6% | 250 mL |
| H2O (tap water) | n/a | 750 mL |
| Total | 1 L |
2. 10% BSA
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| BSA | 10% | 10 g |
| PBS 1× | ||
| Total | 100 mL |
3. PBST
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PBS 10× | 1× | 50 mL |
| Tween 20 | 0.1% | 500 μL |
| H2O (MilliQ water) | n/a | 449.5 mL |
| Total | 500 mL |
4. Methanol dilutions
Make 75%, 50%, and 25% dilutions of methanol in PBST. Prepare the dilutions at least 30 min before the experiment as an exothermic reaction occurs when mixing methanol with PBST and the solution needs to be used at room temperature.
Laboratory supplies
1. Embryo collection cages (Dutscher, catalog number: 789092)
2. Collection plates: 90 mm Petri dishes, half filled with fly culture medium with Neutral Red dye to visualize the eggs. Each plate should be supplemented with 1/3 teaspoon of yeast paste spread on the culture medium
3. Embryo collection basket: cut both the end and the screw cap of a 50 mL Falcon tube and screw the open cap with a square of thin mesh (Dutscher, catalog number: 074007)
4. Medium paint brush (local purchase)
5. Eppendorf 2 mL tube (Dutscher, catalog number: 033297)
6. Pasteur glass pipettes 150 mm (Dutscher, catalog number: 065421)
7. Pipettes 5 mL (Dutscher, catalog number: 357543)
8. Scintillation glass vial (Merck, catalog number: DWK986546)
9. Terumo Agani needle 21G × 1’’ (0.8 mm × 25 mm) (VWR, catalog number: TERUSAN2125R1)
10. StarFrost microscope slides (Dutscher, catalog number: 100204)
11. High-precision microscope cover glasses 22 mm × 22 mm, No.1.5H Marienfeld (VWR, catalog number: 630-2186)
12. Transparent nail polish (local purchase)
Equipment
1. PipetBoy
2. Natural rubber suction aid 2 mL (Roth, catalog number: 1C6P.1)
3. Stereomicroscope (Olympus, model: SZ51)
4. Platform shaker (Heidolph, model: Rotamax 120)
5. Benchtop vortex (Corning, model: LSE Vortex Mixer)
6. Rotating wheel (Snijders, model: 34528 Test Tube Rotator)
7. STED microscope (Abberior)
Software and datasets
1. Imspector (Abberior Instruments)
2. ImageJ/Fiji v2.14.0
3. Microsoft Excel v16.90.2
4. Prism v10 (GraphPad)
5. BioRender (https://www.biorender.com/). The following figures were created using BioRender: Graphical overview: BioRender.com/h93e102; Figure 1: BioRender.com/m96q857; Figure 2: BioRender.com/a28f858.
Procedure
文章信息
稿件历史记录
提交日期: Dec 9, 2024
接收日期: Feb 7, 2025
在线发布日期: Feb 27, 2025
出版日期: Mar 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Haidar, A., Simonelig, M. and Ramat, A. (2025). Visualization of mRNA Translation Within Germ Granule Biphasic Organization in Drosophila Early Embryo. Bio-protocol 15(6): e5242. DOI: 10.21769/BioProtoc.5242.
分类
发育生物学 > 繁殖 > 生殖细胞
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













