- EN - English
- CN - 中文
A Highly Sensitive and Selective DAMP Assay for the Detection of Bacterial Pathogens Associated With Brain Inflammation
高灵敏度、高选择性的 DAMP 检测法用于检测与脑部炎症相关的细菌病原体
发布: 2025年03月20日第15卷第6期 DOI: 10.21769/BioProtoc.5240 浏览次数: 1652
评审: Anupama SinghSrestha DasguptaDebashis Dutta
Abstract
The early detection of meningitis pathogens—including Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae, and Klebsiella pneumoniae—through point-of-care (POC) systems is essential for mitigating the risk of neurological damage, enhancing patient outcomes, and facilitating prompt clinical decision-making. Nucleic acid amplification testing (NAAT) is a promising tool for improving the diagnosis process of bacterial pathogens associated with brain inflammation. This is due to its high sensitivity, rapidity, and compatibility with portable diagnostic platforms, making it particularly suitable for POC applications. This protocol introduces an innovative diagnostic approach designed to function effectively without the need for advanced laboratory equipment. By leveraging dual-priming isothermal amplification (DAMP), the assay uses custom internal primers to enhance specificity and minimize false results. Brilliant Green is used in this assay for fluorescence detection due to its availability, high fluorescence level, and optimal sample-to-background (S/B) ratio. The assay demonstrated excellent specificity, absence of false positives, sensitivity comparable to loop-mediated isothermal amplification (LAMP), and a high S/B ratio.
Key features
• A POC-compatible diagnostic assay developed to enable rapid and accurate diagnosis of meningitis without the need for advanced laboratory infrastructure.
• A DAMP-based approach for the identification of pathogens responsible for bacterial meningitis with high sensitivity and specificity while ensuring the absence of false-positive results.
Keywords: DNA quantitative detection (DNA 定量检测)Graphical overview
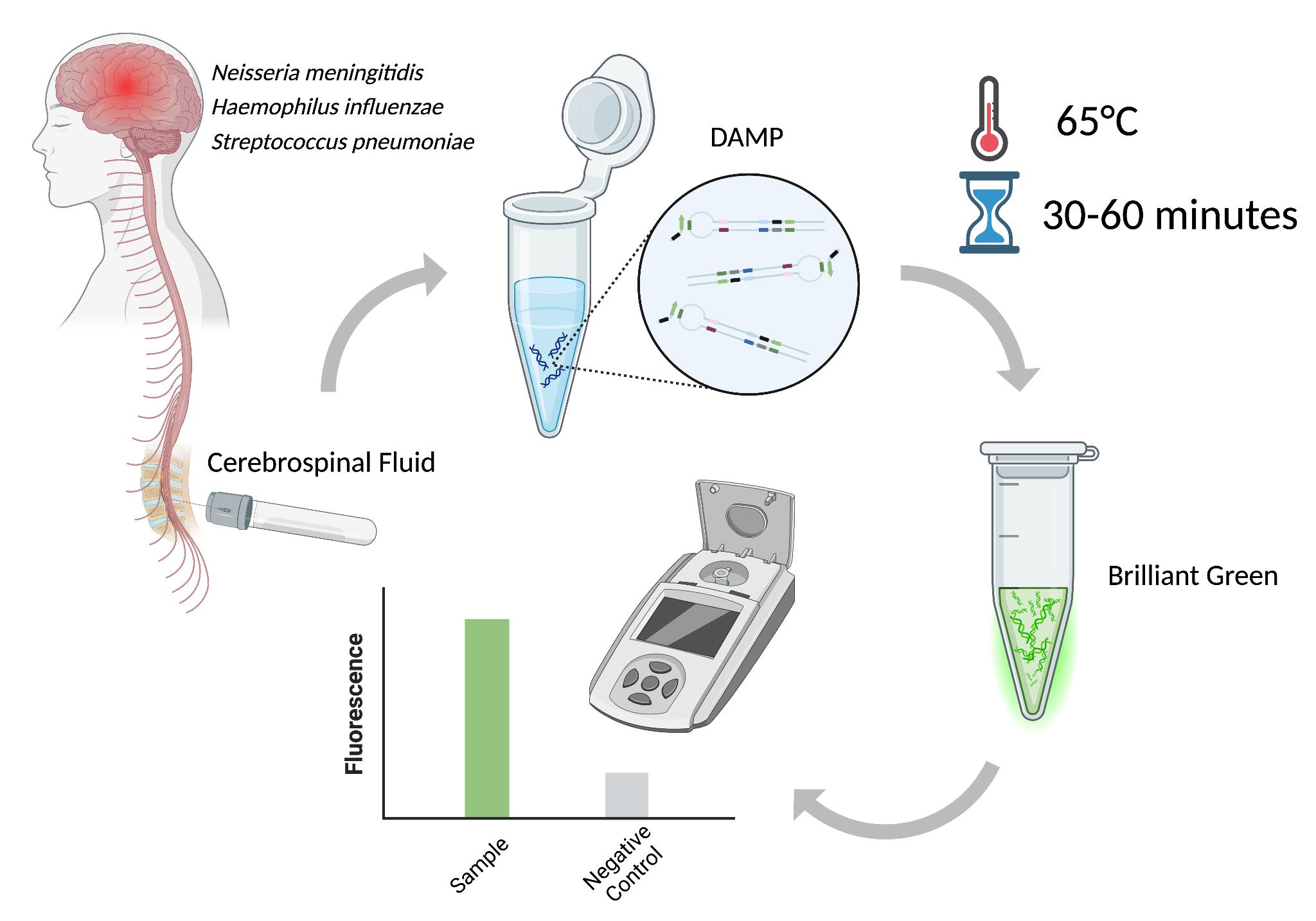
Schematic workflow of this protocol [1]
Background
The rapid and accessible diagnosis of infectious diseases is essential for effective treatment, especially in cases of brain inflammations such as meningitis and encephalitis, where rapid intervention is crucial to preventing complications and decreasing morbidity [2]. For these infections, diagnostic methods need to be highly sensitive, specific, and accessible, ideally matching the point-of-care (POC) parameters: a testing duration of less than 2 h, a cost of less than $10 per test, and suitable for use by untrained personnel. Nucleic acid amplification testing (NAAT) is regarded as the gold standard for molecular diagnostics in this area [3]. PCR-based technologies are among the most commonly used NAAT methods in clinical practice. Recent research indicates that NAATs are essential diagnostic tools [4], particularly in instances where cerebrospinal fluid (CSF) cultures yield negative results [5], as these culturing methods become significantly limited upon antibiotic treatment.
Among NAAT techniques, isothermal amplification has emerged as particularly well-suited for POC diagnostics as it eliminates the need for complex lab equipment [6]. Isothermal amplification eliminates the need for thermal cycling and utilizes specialized enzymes with strand displacement activity to amplify target DNA or RNA sequences at a constant temperature. Of these techniques, loop-mediated isothermal amplification (LAMP) is the most widely used due to its simplicity, rapidity, and high sensitivity. However, despite its advantages, LAMP is prone to nonspecific amplification, which may be acceptable for screening purposes but is unsuitable for guiding therapeutic decisions. Thus, in general practice, LAMP often requires an additional PCR test to confirm primary results [7,8]. A novel method, dual-priming isothermal amplification (DAMP), was developed to overcome the limitations of LAMP. DAMP incorporates a dual-priming mechanism using specialized primers to enhance specificity and reduce false-positive results. While LAMP uses six primers to target eight distinct regions of the target sequence, with inner primers spanning 40–60 nucleotides to facilitate self-priming strand extension, DAMP targets six distinct regions using six primers, with inner primers designed to span less than 40 nucleotides [9]. Additionally, DAMP incorporates pairing-competition primers, which enhance amplification efficiency by balancing self-priming and pairing-priming strand extensions, making it a promising tool for rapid, accurate detection of meningitis in both laboratory and POC settings [9].
A critical aspect of introducing NAAT-based methods into clinical practice is the detection of amplification products. The detection can be achieved through direct or indirect methods. Various visualization techniques can use fluorescent compounds (e.g., DNA intercalating) or naked-eye detection [10]. Although naked eye readouts do not require any specialized tools, their dynamic ranges are often limited, and personnel should have the skills to distinguish subtle changes in turbidity or color. On the other hand, fluorescent detection is preferred for its high sensitivity and quantitative capabilities [11]. Previously, we evaluated three fluorescent dyes for their effectiveness in detecting double-stranded DNA (dsDNA) amplification products: dsGreen, Thioflavin T, and Brilliant Green. Each dye was assessed based on its sample-to-background (S/B) ratio, which is crucial for minimizing background noise and ensuring accurate detection [1]. Brilliant Green was identified as the optimal dye due to its high fluorescence intensity, affordability, and superior S/B ratio. The performance of this dye, combined with the DAMP reaction, provides a highly sensitive and specific assay for detecting the four primary bacterial pathogens responsible for meningitis: Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae, and Klebsiella pneumoniae. These pathogens were selected as the leading causes of bacterial meningitis in individuals beyond the neonatal stage.
The developed assay was evaluated using bacterial culture and clinical cerebrospinal fluid (CSF) samples. It demonstrated both high specificity and sensitivity. Its versatility allows it to function as a standalone diagnostic tool or to be integrated into POC devices, offering a reliable, rapid, and accessible solution for the diagnosis of meningitis.
Materials and reagents
Biological materials
Thermally inactivated cell cultures of the pathogens Klebsiella pneumoniae, Haemophilus influenzae, Neisseria meningitidis, and Streptococcus pneumoniae were obtained from the Pediatric Research and Clinical Center for Infectious Diseases (PRCCID) under the Russian Federal Medical Biological Agency.
Note: This protocol can also be performed on DNA isolated from CSF samples. Thermally inactivated CSF clinical samples were also provided by the PRCCID.
Reagents
1. Nuclease-free water (Invitrogen, catalog number: 1097704)
2. dNTPs (Evrogen, catalog number: PB032)
3. Fine powder MgSO4 (AppliChem, catalog number: 141404)
4. Ethylene glycol liquid (Lenreactiv, catalog number: 263059)
5. Fine-powder KCl (AppliChem, catalog number: 131494.1210)
6. Fine-powder (NH4)2SO4 (AppliChem, catalog number: A3485,5000)
7. Fine powder Tris-HCl (AppliChem, catalog number: 633654.0415)
8. Tween 20% (AppliChem, catalog number: A4974,0500)
9. Bst DNA polymerase (BioLabMix, catalog number: E-10010)
10. Fine powder Brilliant Green (Lenreactiv, catalog number: 020264)
11. 50 bp+ DNA ladder (Evrogen, catalog number: NL003)
12. Ethidium bromide (BioLabMix, catalog number: EtBr-10)
13. 50× TAE buffer (BioLabMix, catalog number: BE-DNA-1000)
14. Fine powder agarose (Helicon, catalog number: SYS-Q0009-0.1)
15. 4× gel loading dye (Evrogen, catalog number: PB020)
16. Primers (Evrogen, direct order):
For Klebsiella pneumoniae:
Outer forward primer (FO): GCGAAGAAACAGAACGGC
Outer reverse primer (RO): TGATCGGGGTCATCTTGC
Inner forward primer (FI): TACGGTCGGAGCTGGTGTAGTTTTCTATGATTTCGGCGGCAG
Inner reverse primer (RI): GACCGGCCTGAAATATGACGCTTTTTACATGGTCGCCAGGTAG
Paring-competition forward primer (FC): TACGGTCGGAGCTGGTGTAG
Pairing-competition reverse primer (RC): GACCGGCCTGAAATATGACGC
Note: The primer sequences for the other pathogens can be found in the original article [11].
Solutions
1. 1× TAE buffer (see Recipes)
2. 10× Bst buffer (see Recipes)
3. Brilliant Green 100 μM stock (see Recipes)
4. MgSO4 1 M stock (see Recipes)
Recipes
1. 1× TAE buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 50× TAE buffer | 1× | 40 mL |
| Deionized water | n/a | 1,960 mL |
| Total | 2,000 mL |
2. 10 × Bst buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| (NH4)2SO4 1 M | 100 mM | 1 mL |
| KCl 1 M | 120 mM | 1 mL |
| MgSO4 1 M (Recipe 4) | 20 mM | 0.2 mL |
| Tris-HCl (pH 8.8) 1 M | 200 mM | 2 mL |
| Tween 20% | 2% | 0.2 mL |
| Ethylene glycol | 5% | 0.5 mL |
| Total | 10 mL |
3. Brilliant Green 100 μM stock
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Brilliant Green | 100 μM | 48.3 ng |
| Deionized water | n/a | 1 mL |
| Total | 1 mL |
4. MgSO4 1 M stock
| Reagent | Final concentration | Quantity or Volume |
| Fine powder MgSO4 | 1 M | 246.47 ng |
| Deionized water | n/a | 1 mL |
| Total | 1 mL |
Laboratory supplies
1. Costar 96-well black polystyrene plate (Corning, catalog number: COS3915)
2. 1–10 μL pipette (Kirgen, catalog number: KG-Pro10)
3. 2–20 μL pipette (Kirgen, catalog number: KG-Pro20)
4. 20–200 μL pipette (Kirgen, catalog number: KG-Pro200)
5. 100–1,000 µL pipette (Kirgen, catalog number: KG-Pro1000)
Equipment
1. Wide mini-sub cell GT Cell (Bio-Rad, catalog number: 1704468EDU)
2. PowerPacTM basic power supply (Bio-Rad, catalog number: 1645050)
3. Spark multimode microplate reader (Tecan, model: Spark-10M)
4. Thermocycler (DNK-Technology, model: DTlite 4S1)
5. ChemiDocTM touch gel imaging system (Bio-Rad, catalog number: 1708370)
Software and datasets
1. Basic Local Alignment Search Tool (BLAST) available on the National Center for Biotechnology Information (NCBI) platform to analyze the sequence data
2. LAMP primer design tool provided by New England Biolabs (NEB)
3. GraphPad Prism v. 9.3.1.
Procedure
文章信息
稿件历史记录
提交日期: Nov 5, 2024
接收日期: Feb 4, 2025
在线发布日期: Feb 27, 2025
出版日期: Mar 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Shkodenko, L. A., Ateiah, M., Mohamed, A., Rubel, M. S. and Koshel, E. I. (2025). A Highly Sensitive and Selective DAMP Assay for the Detection of Bacterial Pathogens Associated With Brain Inflammation. Bio-protocol 15(6): e5240. DOI: 10.21769/BioProtoc.5240.
分类
微生物学 > 病原体检测 > 生物传感器
分子生物学 > DNA > DNA 定量
神经科学 > 神经系统疾病
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













