- EN - English
- CN - 中文
Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells
高灵敏且可调控的 ATOM 荧光生物传感器:用于检测细胞中蛋白质靶点的亚细胞定位
发布: 2025年03月20日第15卷第6期 DOI: 10.21769/BioProtoc.5239 浏览次数: 2270
评审: Xiaokang WuJose Martinez HernandezAnonymous reviewer(s)
Abstract
Fluorescent protein biosensors (FPBs) that turn on—go from dark to bright upon binding their ligands—enable the detection of targets in living cells with high sensitivity and spatial localization. Several approaches exist for creating turn-on FPBs, most notably the method that gave rise to the GCaMP family of genetically encoded calcium indicators. However, it remains challenging to modify these sensors to recognize new ligands. We recently developed adaptable turn-on maturation (ATOM) biosensors, in which target recognition by a small binding domain triggers chromophore maturation in the fluorescent protein to which it is attached. ATOM sensors are advantageous because they are generalizable (by virtue of the monobody and nanobody binding domains) and modular (binding domains and fluorescent proteins of various colors can be mixed and matched for multiplexed imaging), capable of detecting endogenously expressed proteins, and able to function in subcellular compartments including the cytoplasm, nucleus, endoplasmic reticulum, and mitochondria. The protocols herein detail how to design, clone, and screen new ATOM sensors for detecting targets of choice. The starting materials are the genes encoding for a monobody or nanobody and for a cyan, yellow, or red fluorescent protein. We also present general guidelines for creating ATOM sensors using binding domains other than nanobodies and monobodies.
Key features
• Creation of six-member (monobody-based) and nine-member (nanobody-based) plasmid libraries encoding ATOM biosensors.
• Targeting ATOM biosensors to subcellular compartments using peptide tags.
• Screening for biosensor activity in human cells and quantifying turn-on using a fluorescence microscope and freeware software packages.
• The most time-consuming step is ATOM gene construction, which can be bypassed by commercial gene synthesis.
Keywords: ATOM (ATOM)Graphical overview
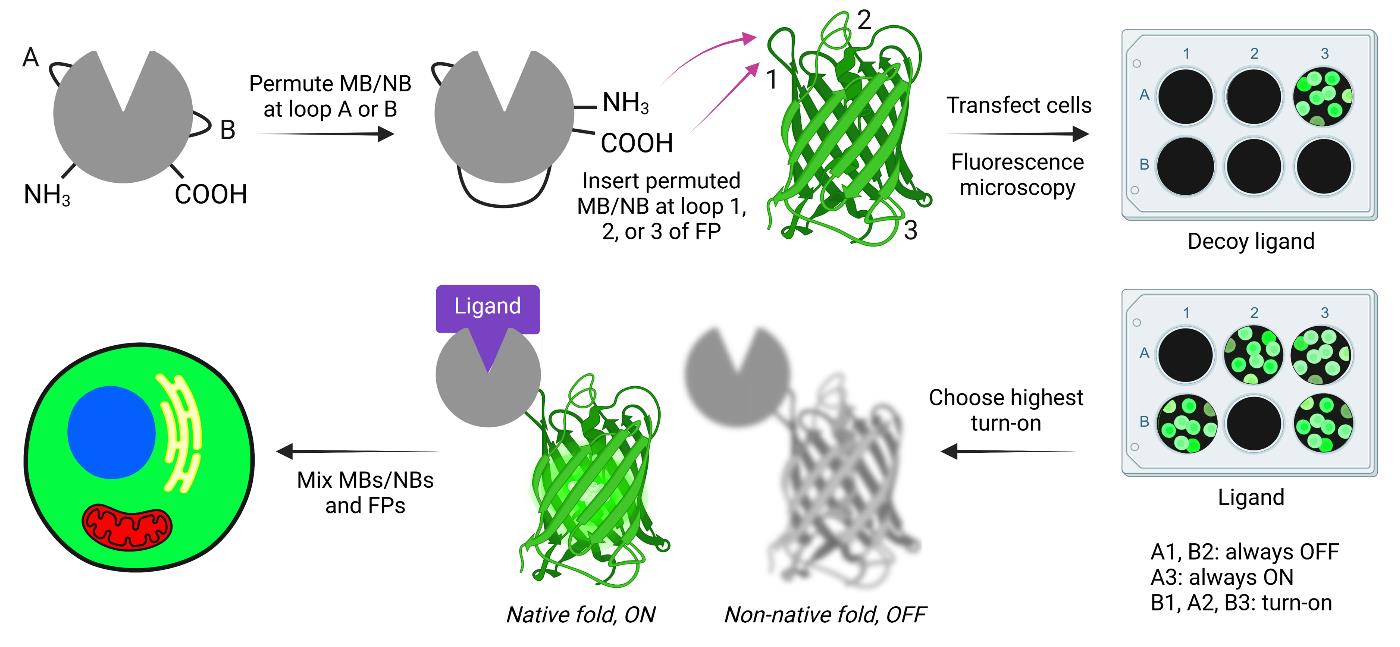
Creating and screening adaptable turn-on maturation (ATOM) biosensors. MB, NB, and FP indicate monobody, nanobody, and fluorescent protein, respectively.
Background
Fluorescent protein biosensors (FPBs) consist of a ligand binding domain coupled to a fluorescent protein (FP), such that binding to the former causes the latter to change the color and/or intensity of its fluorescence. Traditionally, there have been two main mechanisms by which FPBs operate: 1) ligand binding causes a change in the physicochemical environment of the mature chromophore (consisting of three consecutive amino acids that have undergone cyclization and oxidation), or 2) ligand binding either prevents the biosensor from degradation or activates transcription of the biosensor. We recently described a third mechanism, adaptable turn-on maturation (ATOM), in which ligand binding triggers chromophore maturation.
ATOM biosensors are constructed by inserting a circularly permuted binding domain into a surface loop or turn of a fluorescent protein (FP). The amino-to-carboxy terminal distance of most binding domains is too long to be compatible with insertion into surface turns without causing the FP and binding domains to undergo mutually exclusive folding when fused [1]. The binding domains must therefore be circularly permuted first to reduce the distance between their termini. Permutation also serves the purpose of destabilizing the binding domain, which is likely important for the ATOM mechanism. The FP is held in a conformation that does not allow the maturation of its chromophore. Ligand binding then facilitates the folding of the binding domain and FP domain, resulting in chromophore maturation and irreversible intensiometric turn-on.
A key feature of ATOM is that the sensing mechanism is modular and appears to be general. Two different binding domains—monobodies (MBs; ~11 kDa) and nanobodies (NBs; ~14 kDa)—have been shown to be compatible with ATOM [2]. Composed of antiparallel beta strands connected by loops and turns, MBs and NBs recognize ligands via three complementarity-determining loops, leaving the remaining turns (two for MBs and three for NBs) available for circular permutation and fusion into the FP (see section B). MBs and NBs can be adapted to recognize a variety of ligands by in vitro and in vivo selection methods, respectively. We demonstrated generalizability by making ATOM biosensors for four protein targets (WDR5, hRAS, SH2, and mCherry, including the nonfluorescent F70A mCherry variant) with FPs of three colors (mTurquoise, mClover, and mTagRFP). ATOMs function well in the cytoplasm, nucleus, mitochondria, and endoplasmic reticulum [2].
The protocols in this paper detail the steps required to build ATOM biosensors by joining MBs and NBs to FPs of various colors. We also include general considerations if the reader wishes to create ATOM sensors using binding domains other than MBs and NBs (see General Notes 1–3).
Materials and reagents
Biological materials
1. TOP10 chemically competent E. coli cells (Thermo Fisher Scientific, catalog number: C404010)
2. HEK 239T human epithelial-like cells (ATCC, catalog number: CRL-3216)
3. HeLa human epithelial cells (ATCC, catalog number: CCL-2)
4. pCMV-MBP-yATOM_SH2 for expressing y-ATOMSH2 biosensor in mammalian cells (Addgene plasmid # 209708; http://n2t.net/addgene:209708; RRID: Addgene_209708)
5. pCMV-MBP-yATOM_WDR5 for expressing y-ATOMWDR5 biosensor in mammalian cells (Addgene plasmid # 209706; http://n2t.net/addgene:209706; RRID: Addgene_209706)
6. pCMV-MBP-yATOM_mCherry for expressing y-ATOMmCh biosensor in mammalian cells (Addgene plasmid # 209707; http://n2t.net/addgene:209707; RRID: Addgene_209707)
7. pCMV-MBP-yATOM_RAS for expressing y-ATOMRAS biosensor in mammalian cells (Addgene plasmid # 209709; http://n2t.net/addgene:209709; RRID: Addgene_209709)
8. pCMV-RBP_WDR5 for expressing WDR5 in mammalian cells (Addgene plasmid # 209702; http://n2t.net/addgene:209702; RRID: Addgene_209702)
9. pCMV-mCherry (Y70A) for expressing the Y70A (nonfluorescent) mutant of mCherry in mammalian cells (Addgene plasmid # 209703; http://n2t.net/addgene:209703; RRID: Addgene_209703)
10. pCMV-SH2 for expressing SH2 domain in mammalian cells (Addgene plasmid # 209704; http://n2t.net/addgene:209704; RRID: Addgene_209704)
11. PCMV-hRAS (G12V) for expressing hRAS G12V in mammalian cells (Addgene plasmid # 209705; http://n2t.net/addgene:209705; RRID: Addgene_209705)
Reagents
1. Rabbit anti-GFP polyclonal antibody, Alexa Fluor594 (Thermo Fisher Scientific, catalog number: A21312)
2. Rabbit anti-tagRFP polyclonal antibody (Thermo Fisher Scientific, catalog number: R10367)
3. Goat anti-rabbit IgG secondary antibody, Alexa Fluor488 (Thermo Fisher Scientific, catalog number: A11008)
4. Agar (BD Difco, catalog number: 214510)
5. AquaPor LE GTAC agarose (National Diagnostic, catalog number: EC-202-500)
6. Blocking solution (with BSA) 5% in tris-buffered saline (TBS) (Thermo Fisher Scientific, catalog number: J62637.AP)
7. Calcium chloride (CaCl2) dihydrate (J.T. Baker, catalog number: 1332-01)
8. Collagen type I, rat tail (Corning, catalog number: 354236)
9. Deoxynucleotide (dNTP) solution mix (New England Biolabs, catalog number: N0447L)
10. DMEM, high glucose, GlutaMAX supplement, pyruvate (Thermo Fisher Scientific, catalog number: 10569010)
11. DMEM, high glucose, HEPES, no phenol red (Thermo Fisher Scientific, catalog number: 21063029)
12. DpnI restriction endonuclease (New England Biolabs, catalog number: R0176S)
13. DreamTaq DNA polymerase (Thermo Fisher Scientific, catalog number: K1082)
14. Fetal bovine serum (FBS), heat-inactivated (Thermo Fisher Scientific, catalog number: A5670801)
15. GeneJET Gel Extraction kit (Thermo Fisher Scientific, catalog number: K0692)
16. GeneJET Plasmid Miniprep kit (Thermo Fisher Scientific, catalog number: K0503)
17. GeneJET PCR Purification kit (Thermo Fisher Scientific, catalog number: K0702)
18. HEPES (Fisher BioReagents, catalog number: BP310)
19. LB broth, Miller (BD Difco, catalog number: 244610)
20. Kanamycin sulfate (MP Biomedicals, catalog number: 0219453105)
21. Monosodium phosphate monohydrate (J.T. Baker, catalog number: 3818)
22. Paraformaldehyde (PFA), 32% w/v aqueous solution (Thermo Scientific Chemicals, catalog number: 047377.9L)
23. Penicillin-streptomycin (pen/strep) (10,000 units/mL) (Thermo Fisher Scientific, catalog number: 15140122)
24. Phusion high-fidelity DNA polymerase (New England Biolabs, catalog number: M0530S)
25. Q5 high-fidelity DNA polymerase (New England Biolabs, catalog number: M0491L)
26. Sodium chloride (NaCl) (VWR BDH Chemicals, catalog number: BDH9286)
Solutions
1. HEK 293T cell growth media (see Recipes), used in steps F1 and F4
2. 2× HBS (see Recipes), used in step F2
3. PBS (see Recipes), used in steps G2, G4, G8, G10, G11, and H2
4. 3% PFA (see Recipes), used in step G3
Recipes
1. HEK 293T cell growth media
| Reagent | Final concentration | Volume |
|---|---|---|
| DMEM | n/a | 500 mL |
| FBS | 9% | 50 mL |
| Pen/strep | 99 units/mL | 5.5 mL |
| Total | n/a | 555.5 mL |
HEK 293T growth media can be stored at 4 °C for several weeks.
2. 2× HBS
| Reagent | Final concentration | Final pH |
|---|---|---|
| HEPES | 50 mM | 7.0 |
| Monosodium phosphate | 1.5 mM | 7.0 |
| NaCl | 140 mM | n/a |
2× HBS should be frozen at -20 °C in single-use aliquots. Once thawed, any HBS not used that day should be discarded.
3. PBS
| Reagent | Final concentration | Final pH |
|---|---|---|
| Monosodium phosphate | 20 mM | 7.4 |
| NaCl | 150 mM | n/a |
PBS can be stored at room temperature indefinitely if sterility is maintained.
4. 3% PFA
| Reagent | Final concentration | Volume |
|---|---|---|
| PFA (32% stock) | 3% | 468 μL |
| PBS | n/a | 4.532 mL |
| Total | n/a | 5 mL |
3% PFA is prepared fresh for each day’s experiments. The 32% PFA stock solution should be stored at -20 °C in single-use aliquots. Ensure that the stock PFA aliquot is completely thawed and resuspended prior to making the working solution. If needed, heat the stock solution to 42 °C and periodically vortex until fully dissolved.
Laboratory supplies
1. 24-well treated tissue culture plates (NEST, catalog number: 702001)
2. 15 mm coverslip (CELLTREAT Scientific Products, catalog number: 229172)
3. Microscope slides (Fisher Scientific, catalog number: 12-550-143)
Equipment
1. Zeiss Axiovert 200 m inverted microscope with a 10×/0.3 Plan-Neofluor objective
2. Zeiss Axioimager Z1 upright microscope with 10×/0.25 A-plan objective
3. Leica SP8 confocal microscope with 63× oil immersion objective
4. Filter cubes for Zeiss microscopes (Table 1)
Table 1. Filter cube specifications for imaging fluorescent proteins and dyes
| Dye | Excitation | Emission | Dichroic | Zeiss Filter # |
|---|---|---|---|---|
| mTurquoise | 436/20 | 480/40 | 455 | 47 |
| mClover | 500/20 | 535/30 | 515 | 46 |
| mTagRFP-t | 545/25 | 605/70 | 570 | 43 |
| Alexa 595 | 560/40 | 630/75 | 585 | 45 |
| Hoechst | 365 | 445/50 | LP 395 | 49 |
Software and datasets
1. Prism v. 10.4 (GraphPad, 10/23/2024)
2. Fiji [3]
3. PyMol v 3.0 (Schrödinger, LLC)
Procedure
文章信息
稿件历史记录
提交日期: Nov 1, 2024
接收日期: Feb 9, 2025
在线发布日期: Feb 26, 2025
出版日期: Mar 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Sekhon, H., Ha, J. H. and Loh, S. N. (2025). Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells. Bio-protocol 15(6): e5239. DOI: 10.21769/BioProtoc.5239.
分类
分子生物学 > 蛋白质 > 检测
生物化学 > 蛋白质 > 荧光
生物工程 > 生物医学工程
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













