- EN - English
- CN - 中文
Computational Cellular Mathematical Model Aids Understanding the cGAS-STING in NSCLC Pathogenicity
计算细胞数学模型助力解析 cGAS-STING 在非小细胞肺癌发病机制中的作用
(*contributed equally to this work) 发布: 2025年03月05日第15卷第5期 DOI: 10.21769/BioProtoc.5223 浏览次数: 2873
评审: Ran ChenXiaokang WuRobert Kiewisz
Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer. According to 2020 reports, globally, 2.2 million cases are reported every year, with the mortality number being as high as 1.8 million patients. To study NSCLC, systems biology offers mathematical modeling as a tool to understand complex pathways and provide insights into the identification of biomarkers and potential therapeutic targets, which aids precision therapy. Mathematical modeling, specifically ordinary differential equations (ODEs), is used to better understand the dynamics of cancer growth and immunological interactions in the tumor microenvironment. This study highlighted the dual role of the cyclic GMP-AMP synthase–stimulator of interferon genes (cGAS/STING) pathway's classical involvement in regulating type 1 interferon (IFN I) and pro-inflammatory responses to promote tumor regression through senescence and apoptosis. Alternative signaling was induced by nuclear factor kappa B (NF-κB), mutated tumor protein p53 (p53), and programmed death-ligand1 (PD-L1), which lead to tumor growth. We identified key regulators in cancer progression by simulating the model and validating it with the following model estimation parameters: local sensitivity analysis, principal component analysis, rate of flow of metabolites, and model reduction. Integration of multiple signaling axes revealed that cGAS-STING, phosphoinositide 3-kinases (PI3K), and Ak strain transforming (AKT) may be potential targets that can be validated for cancer therapy.
Key features
• Procedures for the reconstruction of a robust and steady-state mathematical model with respective analysis in order to provide mechanistic insights.
• The dynamic mathematical model allows an understanding of the multifaceted dual roles of cGAS-STING in NSCLC promotion and inhibition.
• The inherent statistical tool in systems biology provides a novel immunotherapeutic target.
Keywords: MATLAB (MATLAB)Graphical overview
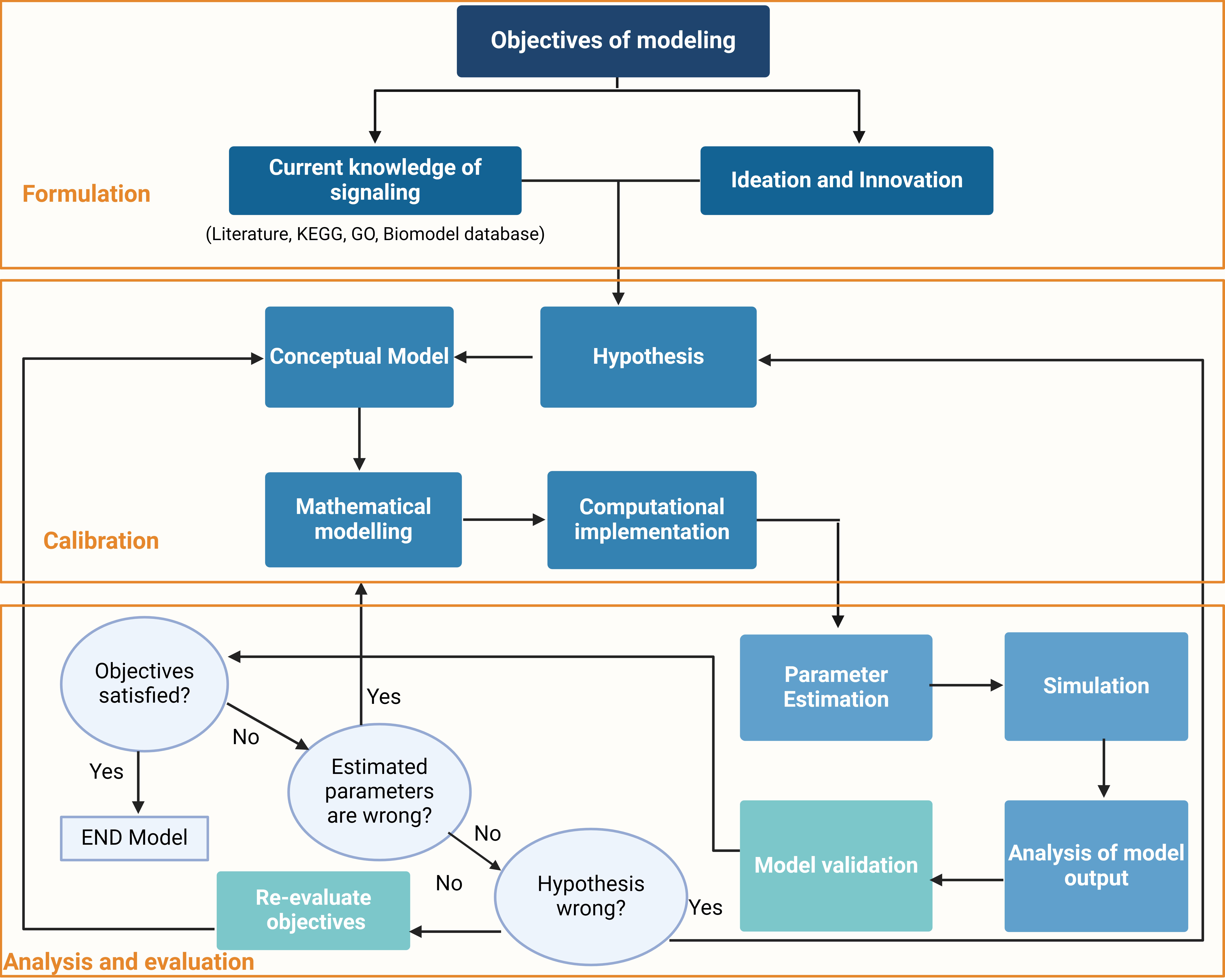
Flowchart of system-based mathematical modeling for the reconstruction of signaling network
Background
Lung cancer is the leading cause of cancer-related deaths worldwide, accounting for approximately 1.79 million fatalities annually [1]. Non-small cell lung cancer (NSCLC) accounts for 85% of cases, with low survival rates with existing treatments. In recent years, NSCLC therapy has undergone an enormous shift since the identification of immunological checkpoints and the subsequent development of immune checkpoint inhibitors (ICIs) [2]. Immunocheckpoint blockade (ICB) targeting the programmed cell death-1/programmed death-ligand1 (PD-1/PD-L1) pathway has become a new standard treatment for NSCLC, improving survival rates either when used alone or in conjunction with conventional chemotherapy [3]. Despite recent developments in immunotherapy, more research is still required to fully comprehend the biology of immune-responsive subsets.
The immune system may be stimulated, and the tumor response to immunotherapy may be improved, by DNA damage, which can be intrinsic due to tumor genomic instability or exacerbated as a result of DNA-damaging treatments [4]. The cGAS-STING signaling pathway is a crucial cytosolic DNA sensing mechanism in vivo, involved in inducing the expression of type I interferons (IFNs) and influencing the body's immune response in controlling pathogen infection, tumor immunity, and autoimmune disorders. cGAS contains an extremely conserved nucleotidyltransferase domain that may detect double-stranded DNA (dsDNA) released into the cytosol [5]. This cGAS-STING signaling in turn triggers IFN I and cytokine response [6–8]. Since aberrant DNA is particularly prominent in cancer as a result of its release from necrotic cells and DNA leakage into the cytosol of cancer cells [9], cGAS signaling may trigger IFN I synthesis, immunological activation, and cancer cell apoptosis. Consequently, downregulating the cGAS-STING pathway would make it possible for cancer cells to avoid immune recognition and response. Furthermore, this pathway's activation can upregulate PD-L1, inhibiting cytotoxic T lymphocyte (CTLs) activity [10]; mutant p53 has also been shown to suppress downstream signaling from cGAS/STING pathway, thereby enabling immune evasion [11].
We studied the cGAS-STING pathway using systems biology, a holistic approach to decode complex, dynamic biological systems with numerous interrelationships and interactions that occur within the confines of time, physiology, and space [12–14]. One crucial tool in systems biology is mathematical modeling, which determines how biological system components interact to produce intricate dynamic behavior. Computational experiments using mathematical models to simulate biological systems could provide valuable insights into the driven mechanisms [15,16]. A unique systems biology toolbox is offered within the MATLAB framework, providing researchers with a versatile platform to explore biological systems, design algorithms, and create simulation tools [17–19]. In this study, we used mathematical modeling to address a knowledge gap by reconstructing the cGAS-STING signaling pathway using MATLAB and understanding its regulatory and molecular drivers through iterative model simulations and data analysis. This included the identification and validation of statistical parameters of the model, such as sensitivity, principal components, and model reduction. Altogether, these studies present a mathematical model as a framework to redesign complex signaling networks; modulating such signaling axes could allow better therapeutic intervention.
Equipment
1. HP Pro 3090MT
Software and datasets
1. MATLAB v7.11.1.866's (SimBiology tool box) (https://www.mathworks.com/products/simbiology.html)
2. COPASI 4.22 (https://copasi.org/)
3. Sigmaplot 10.0 (https://sigmaplot.en.softonic.com/)
4. Microsoft Excel (https://www.microsoft.com/en-us/microsoft-365/previous-versions/microsoft-office-2013)
Procedure
文章信息
稿件历史记录
提交日期: Nov 12, 2024
接收日期: Jan 19, 2025
在线发布日期: Feb 10, 2025
出版日期: Mar 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Khandibharad, S., Gulhane, P. and Singh, S. (2025). Computational Cellular Mathematical Model Aids Understanding the cGAS-STING in NSCLC Pathogenicity. Bio-protocol 15(5): e5223. DOI: 10.21769/BioProtoc.5223.
分类
生物信息学与计算生物学
系统生物学 > 相互作用组 > 基因网络
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













