- EN - English
- CN - 中文
Streamlined Quantification of Microglial Morphology in Mouse Brains Using 3D Immunofluorescence Analysis
小胶质细胞形态的简化定量分析:基于小鼠大脑3D免疫荧光技术
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5218 浏览次数: 3533
评审: Olga KopachAnonymous reviewer(s)
Abstract
Microglial cells are crucial patrolling immune cells in the brain and pivotal contributors to neuroinflammation during pathogenic or degenerative stress. Microglia exhibit a heterogeneous "dendrite-like" dense morphology that is subject to change depending on inflammatory status. Understanding the association between microglial morphology, reactivity, and neuropathology is key to informing treatment design in diverse neurodegenerative conditions from inherited encephalopathies to traumatic brain injuries. However, existing protocols for microglial morphology analyses lack standardization and are too complex and time-consuming for widescale adoption. Here, we describe a customized pipeline to quantitatively assess intricate microglial architecture in three dimensions under various conditions. This user-friendly workflow, comprising standard immunofluorescence staining, built-in functions of standard microscopy image analysis software, and custom Python scripts for data analysis, allows the measurement of important morphological parameters such as soma and dendrite volumes and branching levels for users of all skill levels. Overall, this protocol aims to simplify the quantification of the continuum of microglial pathogenic morphologies in biological and pharmacological studies, toward standardization of microglial morphometrics and improved inter-study comparability.
Key features
• Comparison of 3D microglial architecture between physiological and pathological conditions.
• Quantitative assessment of critical microglial morphological features, including soma volume, dendrite volume, branch level, and filament length.
• Simplified, semi-automated data export and analysis through simple Python scripts.
Keywords: Microglia (小胶质细胞)Graphical overview
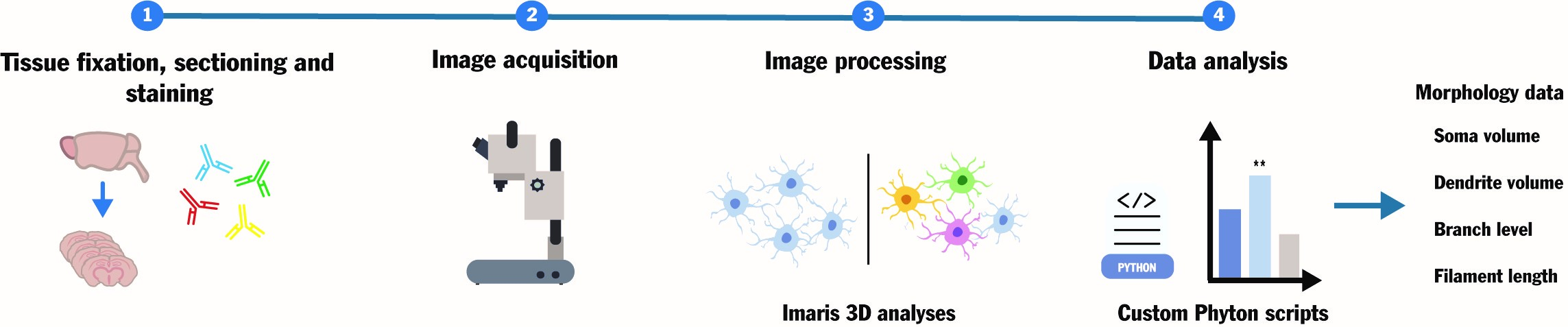
Background
Neuroinflammation, a main mechanism behind neuronal death and dysfunction in various pathologies, involves microglia as key players patrolling immune cells [1]. These cells, known for their morphological heterogeneity and dynamic processes, play diverse roles in different neurodegenerative disorders [2,3]. It is well-known that the functional and morphological alterations in response to intracellular or extracellular cues are highly interconnected. For example, microglia often exhibit increased volume and altered dendrite architecture when performing phagocytic functions in damaged loci, as opposed to a ramified, homeostatic morphology in healthy brain tissue [4,5].
Given the fundamental role of microglia as resident immune cells in the central nervous system, understanding their behavior in pathological contexts is crucial. In this regard, light microscopy–based immunohistochemical (IHC) analyses of ex vivo brain sections remain a core tool for neuropathology research centers worldwide. However, the dense and intricate microglial architecture poses challenges for 3D morphological studies, necessitating a workflow that builds on standard techniques and tools to facilitate standardization and widespread adoption in neuropathology research. Previous methods for 3D IHC analyses have inherent limitations that result in high error rates when measuring numerous small parameters in large and complex brain samples. This often requires significant time investments, specialized analyses, and multiple software tools. Moreover, commonly used techniques for quantitative microglial assessment lack data standardization, resulting in non-comparable results [5]. This is exemplified by the inherent limitations of common open-source tools, such as ImageJ, which either lack the capability for 3D assessment or require complex pipelines involving non-standardized plug-ins to obtain 3D data [3,6–9].
To overcome these challenges and capture the continuum of microglial morphologies under various physiological and pathological states, we describe a straightforward and repeatable custom approach for 3D microglial morphology analyses in mouse brain sections. Our pipeline is based on 3D image acquisition workflows built into the standard Imaris software, followed by custom Python scripts for data analysis. Similar approaches have been successfully employed for 3D analyses of cell volumes and morphological features, albeit in distinct research fields from neuroscience [10,11]. The protocol described here was used to study neuroinflammation and treatment response in primary mitochondrial disorders, specifically in a mouse model of Leigh syndrome that presents distinct patterns of inflammatory neurodegeneration associated with treatment-resistant epilepsy [12,13].
This workflow may be applicable to multiple conditions, providing biological plausibility for the association between gene disruptions and neuropathological phenotypes. It also enables the assessment of the effects of various pharmacological treatments, thereby informing pre-clinical drug development.
Materials and reagents
Biological materials
1. Mice with conditional Ndusf4 ablation in Gad2-espressing neurons (Gad2Cre/+:Ndufs4lox/lox or Gad2:Ndufs4cKO) were obtained as previously described [12,13]. Gad2Cre/+:Ndufs4lox/+ healthy males, expressing Cre specifically in Gad2-positive cells, were crossed with healthy females that have floxed Ndufs4 exon 2 (Gad2+/+, Ndufs4lox/lox), resulting in Gad2:Ndufs4cKO pups or sex/age-matched controls lacking Cre-expression (Gad2+/+) or functional loxP pair on the target gene (Ndufs4lox/+). A full description of the analyses and results pertaining to these samples is described in [13].
Reagents
1. Ketamine (UAB, catalog number: 579557)
2. Sodium chloride (NaCl) (Acros Organics, catalog number: 207790010)
3. Xylazine (Bayer, catalog number: 572126)
4. Phosphate buffered saline (PBS) (Millipore, catalog number: 524650)
5. Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 441244)
6. Ethylene glycol (Merck, catalog number: 102466)
7. Glycerol (Fisher Scientific, catalog number: 12144481)
8. Sodium phosphate (NaH2PO4) (Merck, catalog number: S5011)
9. Triton X-100 (Sigma-Aldrich, catalog number: X100)
10. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: 810533)
11. DAPI-Fluoromount-G solution (Electron Microscopy Science, catalog number: 17984-24)
12. Primary antibody, IBA1 (Wako, catalog number: 019-19741)
13. Secondary antibody, Alexa Fluor 647 (Fisher Scientific, catalog number: A-31573)
Solutions
1. Anesthesia solution (see Recipes)
2. Anti-freezing solution (see Recipes)
3. Permeabilization solution (see Recipes)
4. Blocking solution (see Recipes)
5. Antibody solution (see Recipes)
Recipes
1. Anesthesia solution
100 mg/mL ketamine
20 mg/mL xylazine
0.9% NaCl
2. Anti-freezing solution
30% ethylene glycol
30% glycerol
0.1 M sodium phosphate
3. Permeabilization solution
0.2% Triton X-100
0.1 M PBS
4. Blocking solution
3% BSA
0.1 M PBS
5. Antibody solution
1% BSA
0.15% Triton X-100
0.1 M PBS
Laboratory supplies
1. 24-well plates (Fisher Scientific, catalog number: 11835275)
2. Slides (Fisher Scientific, catalog number: J1800AMNZ)
3. Cover glasses (VWR, catalog number: 631-1339)
Equipment
1. Confocal microscope (Zeiss, model: LSM780)
2. Peristaltic pump for mouse perfusion (Gilson Inc, model: Gilson Minipuls 2)
3. Vibratome (Leica, model: VT 1000S Vibrating-blade microtome)
Software and datasets
1. Imaris software (Bitplane, Belfast, UK, v.9.5, October 17, 2019) with modules for data management, visualization, 3D rendering, and analysis
2. Zeiss LSM Image Browser (version LSM780, 2010)
a. Python 3.11.2 (February 8, 2023)
b. Microsoft Excel (Microsoft, Redmon, WA, Office 365, April 2023)
Code and software availability
Imaris software can be obtained with a license key from the manufacturer (BitPlane, under Oxford Instruments, Belfast, UK). The Python scripts developed here are available from GitHub: https://github.com/AndreudeDonato/Read_Imaris_zstack.git
Data availability
All data shown here are from Puighermanal et al. [13] and are available online at https://doi.org/10.1038/s41467-024-51884-8.
Procedure
文章信息
稿件历史记录
提交日期: Oct 16, 2024
接收日期: Jan 8, 2025
在线发布日期: Jan 26, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
de Donato, M. H., Kouchaeknejad, A., de Donato, A., Van Der Walt, G. and Puighermanal, E. (2025). Streamlined Quantification of Microglial Morphology in Mouse Brains Using 3D Immunofluorescence Analysis. Bio-protocol 15(4): e5218. DOI: 10.21769/BioProtoc.5218.
分类
生物信息学与计算生物学
神经科学 > 细胞机理 > 小神经胶质
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












