- EN - English
- CN - 中文
A Novel Gene Stacking Method in Plant Transformation Utilizing Split Selectable Markers
利用分裂可选择标记的植物转化新型基因叠加方法
(*contributed equally to this work) 发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5214 浏览次数: 1953
评审: Xiaofei LiangHao ChenAnonymous reviewer(s)
Abstract
Gene stacking, the process of introducing multiple genes into a single plant to enhance desired traits, is essential for plant genetic improvement through both conventional breeding and genetic transformation. In general, transformation-based gene stacking can be achieved through either co-transformation to simultaneously introduce multiple genes or sequential multi-round transformation. While co-transformation is generally faster and more efficient than sequential multi-round transformation, it often requires two selectable marker genes, which confer resistance to antibiotics, for selecting transgenic events. However, in most cases, there is only one best selectable marker gene for a specific plant species or genotype. Also, it is harder to optimize the concentrations of two antibiotics for co-transformation than using one antibiotic for selecting transgenic events. To overcome this challenge, we recently developed an innovative split selectable marker system for plant co-transformation, allowing the use of one selectable marker gene to select transgenic events. This method involves constructing two binary vectors, each carrying a subset of genes of interest and a partial fragment of the selectable marker gene, which is connected to a partial intein fragment. Following Agrobacterium-mediated co-transformation, plants harboring both binary vectors are selected using a single antibiotic, such as kanamycin. This split-marker system can be used to co-transform multiple genes into both herbaceous and woody plants, accelerating genetic improvement of polygenic traits or integrative improvement of multiple traits to simultaneously increase crop yield and quality.
Key features
• Developed an intein-mediated split selectable marker system for efficient gene stacking in plants.
• Utilizes a single antibiotic for identifying transgenic events, simplifying the selection process of co-transformation compared to traditional methods.
• Applicable to both herbaceous and woody plant species for co-transforming multiple genes.
• Enhances scalability and feasibility of gene stacking in plant genetic engineering and crop improvement initiatives.
Keywords: Intein (内蛋白)Graphical overview
Background
Gene stacking is a pivotal strategy in modern agriculture, involving the integration of multiple beneficial genes into a single organism to simultaneously enhance traits such as yield, disease resistance, stress tolerance, and nutritional content [1]. This approach is particularly valuable in the metabolic engineering of plants due to the intricate nature of metabolic processes and biochemical pathways, which typically involve complex interactions among multiple genes [2]. Gene stacking in plants can be achieved through several methods. First, hybrid stacking involves cross-hybridizing a plant containing one or more transgenes with another plant carrying different transgenes, resulting in the development of a multi-stack hybrid through iterative hybridization. Second, co-transformation entails transforming a plant with two or more independent transgenes, each in separate DNA constructs delivered simultaneously to the plant. Third, sequential multi-round transformation involves re-transformation of a plant already harboring a transgene with additional transgenes [3].
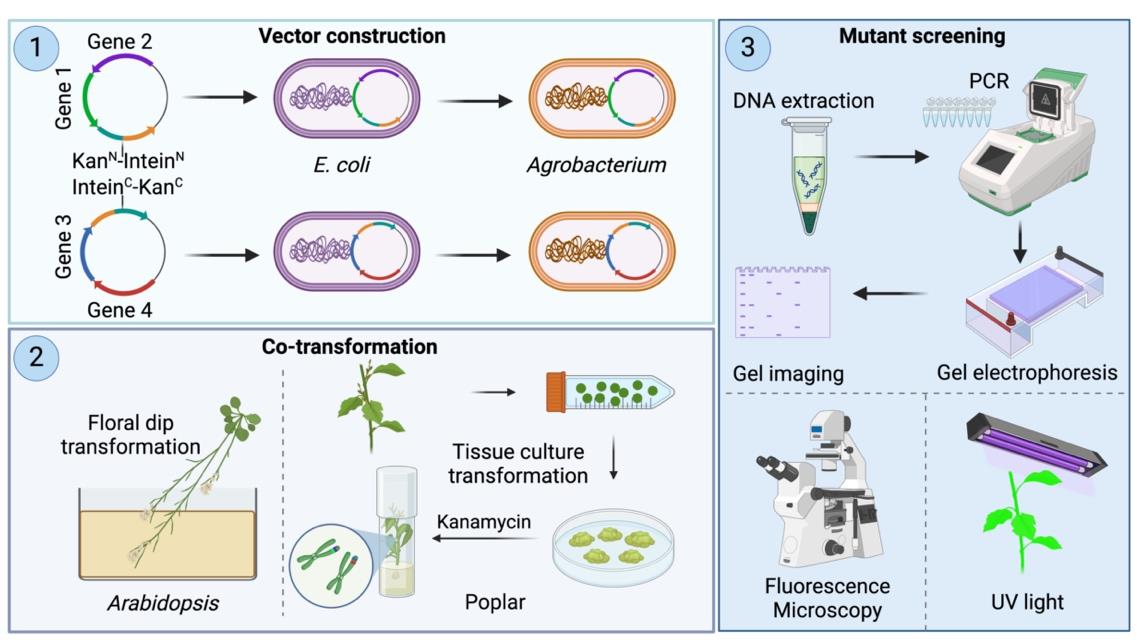
Co-transformation has proven to be a highly promising approach for introducing multiple genes into plants [4]. Typically, co-transformation involves using two different selectable marker genes simultaneously, which must be compatible with both the plant species and the transformation method employed. Challenges arise in ensuring these markers are effective and efficient, requiring careful optimization of transformation protocols and selection conditions to maintain high transformation efficiency. Only a limited number of marker genes, such as hygromycin phosphotransferase (hpt), neomycin phosphotransferase II (nptII), and bar, are commonly used in plant research and crop development, and no single selectable marker gene has been found to be universally effective in all situations [5,6]. To address this challenge, we recently used a split selectable marker system to facilitate gene stacking in plants [7]. This system contains two independent binary vectors, each carrying distinct genes of interest. Unlike the traditional co-transformation system, in which each vector typically contains a different selectable marker, each vector in our split selectable marker system contains a partial fragment of the selectable marker gene, which is connected to a partial intein fragment. The partial intein fragment refers to a segment of an intein that facilitates protein splicing in engineered systems. After the two vectors are co-transformed into the same plant cell, intein-mediated protein splicing occurs post-translationally, resulting in the reassembly of two marker protein fragments into a full-length selectable marker protein. In this natural process, an intein, a protein segment within a precursor protein, self-excises and ligates the remaining protein fragments (exteins) to form a functional protein [8]. This innovative system simplifies the identification of transgenic events generated by co-transformation using a single selection agent (e.g., antibiotic). This approach avoids the inefficiencies and incompatibilities associated with using two different selectable markers in the traditional co-transformation. Designing and constructing intein-mediated selectable markers is highly relevant to molecular cloning. Here, we describe in great detail the methodology—used in the original publication by Yuan et al. [7]—for creating split-marker constructs, co-transforming two vectors in two plant species (Arabidopsis thaliana and Populus tremula × P. alba clone INRA 717-1B4), and confirming transgenic events. Also, we created ready-to-use binary vectors to simplify the cloning procedure for creating DNA constructs for co-transformation.
Materials and reagents
Reagents
1. Agar (PhytoTech LABS, catalog number: HWW1000003C)
2. Agarose (GenBiotech, catalog number: RU1010)
3. Acetosyringone (Sigma-Aldrich, catalog number: D134406)
4. Cefotaxime (PhytoTech LABS, catalog number: C380)
5. NEBridge® Golden Gate Assembly kit (BsaI-HF® v2) (NEB, catalog number: E1601L)
6. NEB® 5-alpha competent E. coli (NEB, catalog number: C2987H)
7. PCR Purification kit (Inbio Highway, catalog number: K1206)
8. EHA105 Agrobacterium ElectroCompetent cells
9. Plasmid DNA Purification kit (Qiagen, catalog number: 12123)
10. Restriction enzymes: Depending on the restriction sites on the inserts that the user cloned
11. Sterilizing agents: ethanol
12. Silwet L-77
13. GoTaq® G2 Master Mixes (Promega, catalog number: 0000434787)
14. Murashige & Skoog basal medium with vitamins (MS) (PhytoTech LABS, catalog number: M5531)
15. MES (Sigma, catalog number: SLCF3242)
16. NAA (Sigma, catalog number: RNBJ597610)
17. IBA (PhytoTech LABS, catalog number: I538)
18. BAP (PhytoTech LABS, catalog number: B130)
19. 2iP; 6-(γ,γ-Dimethylallylamino)purine Solution (PhytoTech LABS, catalog number: D217)
20. 0.5 M EDTA (Millipore, catalog number: 3740903)
21. 5 M NaCl (Sigma, catalog number: SLBR7232V)
22. 1 M Tris-HCl (Invitrogen, catalog number:15567-027)
23. Chloroform, EMSURE® ACS, ISO, Reag. Ph. (VWR, catalog number: 1.02445.1000)
24. Isopropanol (2-Propanol) (VWR, catalog number: 1.09634.5000)
25. L-Glutamine (PhytoTech LABS, catalog number: HWY0229022A)
26. Phyto agar (Sigma, catalog number: P8169)
27. Kanamycin (PhytoTech LABS, catalog number: HHW47510054)
28. LB broth with agar (Sigma, catalog number: L3147)
29. LB broth (Sigma, catalog number: L3022)
30. Sucrose (Cicarelli, catalog number: 841214)
31. Sterile water
32. Timentin (GoloBio, catalog number: T-104-25)
33. Liquid nitrogen
34. Gel Extraction kit (Zymoclean Gel DNA Recovery kit, catalog number: D4008)
35. Q5® High-Fidelity 2× master mix (NEB, catalog number: M0492S)
36. TE buffer (Thermo Fisher Scientific, catalog number: 12090015)
37. Thidiazuron (TDZ) solution, 1 mg/mL (PhytoTech Labs, catalog number: T7999)
38. Cetyltrimethylammonium bromide (CTAB) (Sigma-Aldrich, catalog number: 219374)
Primers (for genotyping)
eYGFPuv_F: 5'-CACGGCAACCTCAACG-3'
eYGFPuv_R: 5'-CTCGACACGTCTGTGGG-3'
Solutions
1. Dip solution (see Recipes)
2. Callus induction media (CIM) (see Recipes)
3. Shoot induction media (SIM) (see Recipes)
4. Shoot elongation media (SEM) (see Recipes)
5. Root induction media (RM) (see Recipes)
6. LB agar medium (see Recipes)
7. LB medium (see Recipes)
8. MS induction medium (see Recipes)
9. 3% CTAB extraction buffer (see Recipes)
Recipes
1. Dip solution
5% sucrose
0.03% Silwet L-77
120 mL deionized water
2. Callus induction media (CIM)
4.3 g/L MS with vitamins
30 g/L sucrose
3 g/L Phyto agar
0.2 g/L L-glutamine
0.25 g/L MES
1 mL of 10 μM/L NAA
Sterilize and add 2ip (5 μM final) 1 mL of 5 mM 2ip stock, timentin (200 mg/mL stock) 1 mL/L, cefotaxime (300 mg/mL stock) 1 mL/L, and kanamycin at 100 mg/L (final concentration).
3. Shoot induction media (SIM)
4.3 g/L MS with vitamins
30 g/L sucrose
3 g/L Phyto agar
0.2 g/L L-glutamine
0.25 g/L MES
50 μL of 1 mg/mL TDZ
Sterilize and add timentin (200 mg/mL stock) 1 mL/L, cefotaxime (300 mg/mL stock) 1 mL/L, and kanamycin at 100 mg/L (final concentration).
4. Shoot elongation media (SEM)
4.3 g/L MS with vitamins
30 g/L sucrose
3 g/L Phyto agar
0.2 g/L L-glutamine
0.25 g/L MES
100 μL of 1 mg/mL BAP
Sterilize and add timentin (200 mg/mL stock) 1 mL/L, cefotaxime (300 mg/mL stock) 1 mL/L, and kanamycin at 100 mg/L (final concentration).
5. Root induction media (RM)
2.15 g/L MS with vitamins
20 g/L sucrose
3 g/L Phyto agar
0.2 g/L L-glutamine
0.25 g/L MES
100 μL of 1 mg/mL IBA
Sterilize and add timentin (200 mg/mL stock) 1 mL/L, cefotaxime (300 mg/mL stock) 1 mL/L, and kanamycin at 100 mg/L (final concentration).
6. LB agar medium
40 g/L LB broth with agar
7. LB medium
20 g/L LB broth
8. MS induction medium
4.43 g/L MS with vitamins
20 g/L sucrose
0.5 g/L MES
20 μM (final concentration) acetosyringone
9. 3% CTAB extraction buffer (50 mL)
Dissolve 1.5 g of CTAB in 16.5 mL of distilled water on a heating plate.
Add 7.5 mL of 1 M Tris-HCl (pH 7.4), 26 mL of 5 M NaCl, and 0.2 mL of 0.5 M EDTA (pH 8).
Filter-sterilize using a 0.2 μm filter.
Laboratory supplies
1. 200 μL PCR tubes (Sigma, catalog number: Z316121)
2. 1.5 mL microcentrifuge tubes (Sigma, catalog number: SLMTBP15-EP)
3. 50 mL Falcon tubes (VWR, catalog number: 21008-940)
4. 14 mL round-bottom tubes (Sigma, catalog number: Z617806)
5. Plant culture vessel (PhytoCon, 16 oz, 473 mL)
6. Pipette and pipette tips (Eppendorf)
7. 100 mm × 15 mm Petri dishes (Sigma, catalog number: Z666246)
8. Scalpel and forceps
9. Gloves
Equipment
1. E. coli and Agrobacterium pulser transformation apparatus (Bio-Rad, model: 155103)
2. High-speed centrifuges (Thermo Fisher Scientific, SorvallTM ST8 Small Benchtop Centrifuge; catalog number: 75007200)
3. Floor model centrifuge (Thermo Fisher Scientific, SorvallTM ST8 FR Floor-Standing Refrigerated Centrifuge; catalog number: 75007208)
4. Incubator (Sigma, catalog number: Z763330)
5. Incubator shaker (Sigma, model: Innova 44 incubator shaker)
6. Biosafety cabinets (LABCONCO, catalog number: 302380011)
7. Plant growth chamber (NORLAKE Scientific)
8. Autoclave
9. Vortex (FISHER MINI VORTEXER, catalog number: 02215365)
10. Nanodrop (Denovix DS11)
11. Gel Doc (Azure Biosystems 600)
12. Cuvette (Bio-Rad, 0.2 cm electrode gap cuvette, catalog number: 1652086)
Software and datasets
1. SnapGene (GSL Biotech LLC, https://www.snapgene.com/)
Procedure
文章信息
稿件历史记录
提交日期: Sep 3, 2024
接收日期: Jan 5, 2025
在线发布日期: Jan 16, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Yuan, G., Islam, M. T., Tuskan, G. A. and Yang, X. (2025). A Novel Gene Stacking Method in Plant Transformation Utilizing Split Selectable Markers. Bio-protocol 15(4): e5214. DOI: 10.21769/BioProtoc.5214.
分类
植物科学 > 植物转化 > 农杆菌介导的转化方法
植物科学 > 植物分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link