- EN - English
- CN - 中文
Real-time IncuCyte® Assay for the Dynamic Assessment of Live and Dead Cells in 2D Cultures
实时IncuCyte®检测用于动态评估二维培养中活细胞和死细胞
发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5210 浏览次数: 2870
评审: Ralph Thomas BoettcherShun Yu Jasemine YangAnonymous reviewer(s)
Abstract
Cell viability and cytotoxicity assays are commonly used to investigate protein function and to evaluate drug efficacy in cancer and other disease models. Cytotoxicity is the measure of dead or damaged cells and is often quantified using assays based on cellular characteristics such as membrane integrity or mitochondrial metabolism. However, these assays are typically limited to endpoint analysis and lack emulation of physiological conditions. The IncuCyte Live and Dead Cell assay described here leverages common cell permeability methodologies but uses fluorescence microscopy channels to image both live and dead cells over time and phase microscopy channels to measure confluency. Cytotox green reagent is a cell membrane–impermeable dye that can only be taken up by cells with poor cell membrane integrity. NucLight rapid red dye is a cell membrane–permeable nuclear dye that can be taken up by all cells. Based on dye uptake and fluorescence intensity, the IncuCyte software can be used to analyze images for live and dead cell detection and quantification. Phase microscopy is used to determine confluency and can be further quantified using the IncuCyte software. We provide an application of this assay, using it to calculate IC50 and EC50 values for the assessment of drug efficacy.
Key features
• Quantify live and dead cells over time.
• Determine drug IC50 and/or EC50 in 2D cell cultures.
• This protocol requires the instrument IncuCyte® S3 (or SX5) Live-Cell Analysis system and corresponding software.
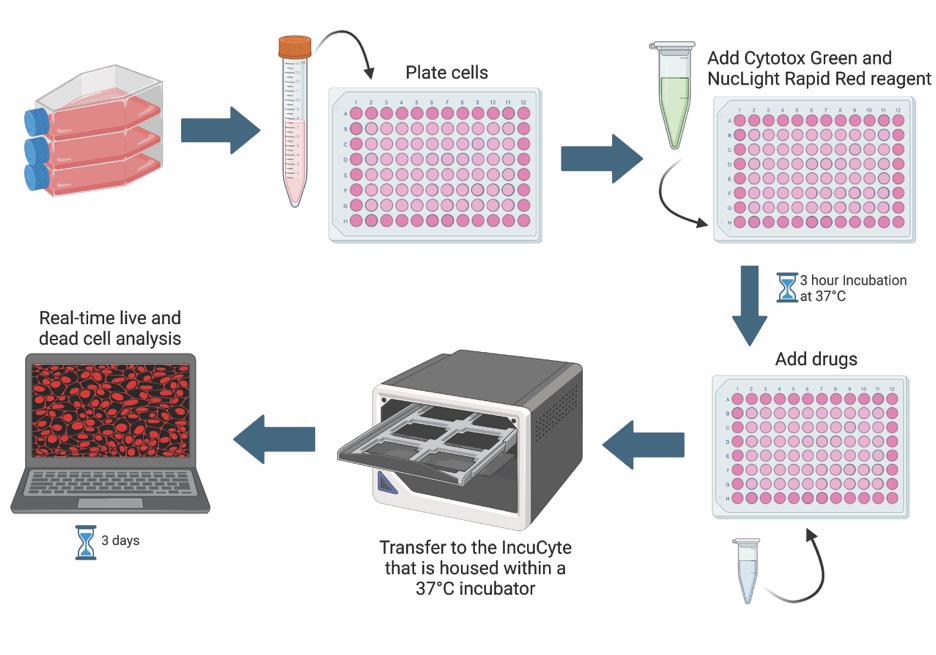
Keywords: IncuCyte (IncuCyte)Graphical overview
Background
Under normal physiological conditions, many cell types undergo turnover to help maintain a balance between cell death and cell survival. However, in cancer contexts, there is an imbalance that favors cell survival or proliferation over cell death. Cancer cell death is an important area of cancer research, especially in drug discovery [1]. High-throughput, sensitive, and robust assays are necessary for screening, testing, and comparing the effectiveness of candidate drugs [1]. The effectiveness of drugs is commonly measured by the half-maximal inhibitory concentration (IC50) or the half-maximal effective concentration (EC50) [2]. The IC50 is the concentration at which half of the measured response is inhibited, while the EC50 is the concentration at which half of the maximum response is induced [2]. One way of measuring drug effectiveness in cancer models is by determining how well a drug induces cytotoxicity [1]. Cytotoxicity can be evaluated via various indirect measures of cell death including cell permeability (trypan blue dye exclusion assay), metabolism (tetrazolium reduction assay), protease activity, ATP detection, and many more [3,4]. In addition to these assays being indirect measures of cell death, other drawbacks typically include the lack of physiological conditions and restriction to end-point analyses [5]. One way to overcome these limitations is by using the IncuCyte Live and Dead Cell assay.
The IncuCyte Live and Dead Cell assay employs common cell permeability techniques, but in a capacity that allows for real-time and live-cell quantification. Cytotox green reagent is a membrane-impermeable dye that is only able to enter cells with poor membrane integrity, hence serving as a cell death marker. NucLight rapid red dye is a membrane-permeable DNA dye that labels all cell nuclei. Residing in a tissue culture incubator, the IncuCyte® Live-Cell Analysis system can capture images at fixed time intervals, enabling the quantification of dead vs. live cells using the IncuCyte software. Live and dead cell quantification is used to calculate the IC50 and/or EC50. Changes in cell morphology, cell migration, and cell–cell clustering interactions throughout the run can also be quantified.
In this protocol, we describe stepwise how to perform the IncuCyte Live and Dead Cell assay, providing an example of how it can be used to determine the IC50 and EC50 of chloroquine (CQ), a late-stage autophagy inhibitor, in the pancreatic ductal adenocarcinoma cell line, MIA PaCa-2. A modified version of this protocol was used in Sathiyaseelan et al. [6].
Materials and reagents
Biological materials
1. MIA Paca-2 cells (ATCC, catalog number: CRL-1740)
Reagents
1. Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, Gibco, catalog number: 11995-065)
2. Fetal bovine serum (FBS) (Thermo Fisher Scientific, Gibco, catalog number: 12483-020)
3. HEPES (Thermo Fisher Scientific, Gibco, catalog number: 15630-080)
4. Insulin solution from bovine pancreas (Sigma-Aldrich, catalog number: 10516)
5. MEM non-essential amino acids (NEAA) (Thermo Fisher Scientific, Gibco, catalog number: 11140-050)
6. 0.25% Trypsin-EDTA phenol red (Thermo Fisher Scientific, Gibco, catalog number: 25200056)
7. PBS, pH 7.4 (Thermo Fisher Scientific, Gibco, catalog number: 10010-049)
8. EVETM slides and 0.4% trypan blue solution (NanoEnTek, catalog number: EVS-050)
9. Chloroquine diphosphate (Sigma-Aldrich, catalog number: C6628-25G)
10. Staurosporine (Sigma-Aldrich, catalog number: S6942)
11. IncuCyteTM NucLight rapid red dye for live-cell nuclear labeling (Sartorius, catalog number: 4717)
12. IncuCyteTM Cytotox green reagent for counting dead cells (Sartorius, catalog number: 4633)
Solutions
1. Full media (see Recipes)
2. Chloroquine (CQ) stock solution (see Recipes)
Recipes
1. Full media (500 mL)
Note: Store the media for up to 2–3 weeks at 4 °C. Warm to 37 °C before using.
| Reagent | Stock concentration | Final concentration | Quantity or Volume |
|---|---|---|---|
| DMEM | n/a | n/a | 439.75 mL |
| FBS | 100% | 10% | 50 mL |
| NEAA | 100× | 1× | 5 mL |
| HEPES | 1 M | 10 mM | 5 mL |
| Insulin | 10 mg/mL | 5 μg/mL | 250 μL |
2. Chloroquine (CQ) stock solution (1.9 mL)
Note: The stock is 100 mM and is stable for at least 1 year at -20 °C. For experiments, dilute to 2 mM (5 µL of 100 mM stock + 245 µL media) in full media and make further dilutions accordingly in full media.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Chloroquine diphosphate (MW: 515.86) | 100 mM | 0.1 g |
| 1× PBS | n/a | 1.9 mL |
Laboratory supplies
1. T75 or T25 flasks (VWR, catalog number: BD353136 or 29185-300)
2. 70% ethanol (HealthCare Plus, catalog number: NPN 00825751)
3. 96-well tissue culture plates (Corning, Falcon, catalog number: 353072)
4. Sterile 15 mL tubes (Corning, Falcon, catalog number: 352096)
5. 10 mL pipette (Corning, catalog number: 4488) and pipet-aid (or aspirator)
6. 5.0 mL screw cap MacroTubes (MTC Bio Incorporated, catalog number: C2540)
7. Syringes for DISTRIMAN Mini ST 1,250 μL (Gilson, catalog number: F164140)
Equipment
1. DISTRIMAN repetitive pipette (Gilson, model: F164001)
2. EVE automatic cell counter (NanoEnTek, catalog number: EVE-MC)
3. Autoflow IR Direct Heat CO2 incubator (NuAire, model: NU-5510E)
4. IncuCyte S3 live-cell analysis system (Sartorius, IncuCyte S3)
5. Tissue culture hood (Microzone Corporation, model: BK-2-4)
6. Vortex Genie 2 (Scientific Industries, model: SI-0236)
Software and datasets
1. IncuCyte software 2022B (IncuCyte, 2022)
2. Excel 2016 (Microsoft, 09/22/2015); requires license
3. Prism v10.2.3 (GraphPad, 04/21/2024); requires license
4. BioRender (https://www.biorender.com/). The following figures were created using BioRender: Graphical overview: BioRender.com/c77h076
Procedure
文章信息
稿件历史记录
提交日期: Aug 30, 2024
接收日期: Dec 30, 2024
在线发布日期: Jan 19, 2025
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gidda, A. K., Chittaranjan, S. and Gorski, S. M. (2025). Real-time IncuCyte® Assay for the Dynamic Assessment of Live and Dead Cells in 2D Cultures. Bio-protocol 15(3): e5210. DOI: 10.21769/BioProtoc.5210.
分类
癌症生物学 > 细胞死亡 > 细胞生物学试验 > 细胞活性
细胞生物学 > 细胞活力 > 细胞死亡
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link