- EN - English
- CN - 中文
Streamlined Quantification of p-γ-H2AX Foci for DNA Damage Analysis in Melanoma and Melanocyte Co-cultures Exposed to FLASH Irradiation Using Automated Image Cytometry
利用自动化图像细胞计数法简化量化p-γ-H2AX焦点分析,以评估暴露于FLASH辐射的小鼠黑色素瘤及黑色素细胞共培养中的DNA损伤
(*contributed equally to this work) 发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5208 浏览次数: 2596
评审: Ismail TahmazIvonne SehringAnonymous reviewer(s)
Abstract
In response to DNA-damaging physical or chemical agents, the DNA damage repair (DDR) pathway is activated in eukaryotic cells. In the radiobiology field, it is important to assess the DNA damage effect of a certain irradiation regime on cancer cells and compare it to the effect on non-transformed cells exposed to identical conditions. The first step in the DNA repair mechanism consists of the attachment of proteins such as the phosphorylated histone γ-H2AX (p-γ-H2AX) to DNA double-strand breaks (DSB) in the nucleus, which leads to the formation of repairing foci. Therefore, imaging methods were established to evaluate the presence of foci inside the nucleus after exposure to DNA-damaging agents. This approach is superior in sensitivity to other methods, such as the comet assay or the pulsed-field gel electrophoresis (PFGE), that allow direct detection of cleaved DNA fragments. These electrophoresis-based methods require high ionizing radiation dosages and are difficult to reproduce compared to imaging-based assays. Conventionally, the number of foci is determined visually, with limited accuracy and throughput. Here, by exploring the effect of laser-plasma accelerated electrons FLASH irradiation on cancer cells, we describe an image cytometry protocol for the quantification of foci with increased throughput, upon large areas, with increased precision and sample-to-sample consistency. It consists of the automatic scanning of fluorescently labeled cells and using a gating strategy similar to flow cytometry to discriminate cells in co-culture based on nuclei elongation properties, followed by automatic quantification of foci number and statistical analysis. The protocol can be used to monitor the kinetics of DNA repair by quantification of p-γ-H2AX at different time points post-exposure or by quantification of other DNA repair proteins that form foci at the DNA DSB sites. Also, the protocol can be used for quantifying the response to chemical agents targeting DNA. This protocol can be performed on any type of cancer cells, and our gating strategy to discriminate cells in co-culture can also be used in other research applications.
Key features
• Analysis of DNA-damage sensitivity using model cancer cell lines and non-transformed cellular controls.
• Allows comparative testing of various doses of DNA damaging radiation on cancer and non-transformed cells in co-culture, as well as in monocultures.
• This protocol requires TissueFAXSiPlus model i12 or an alternative instrument that allows automatic image acquisition and stitching to benefit from enhanced analysis throughput.
• For analyses of co-cultures or heterogeneous samples, TissueQuest software is required to selectively quantify different cell subpopulations; dedicated training is advisable before operating the system.
Keywords: Cancer cells (癌细胞)Graphical overview
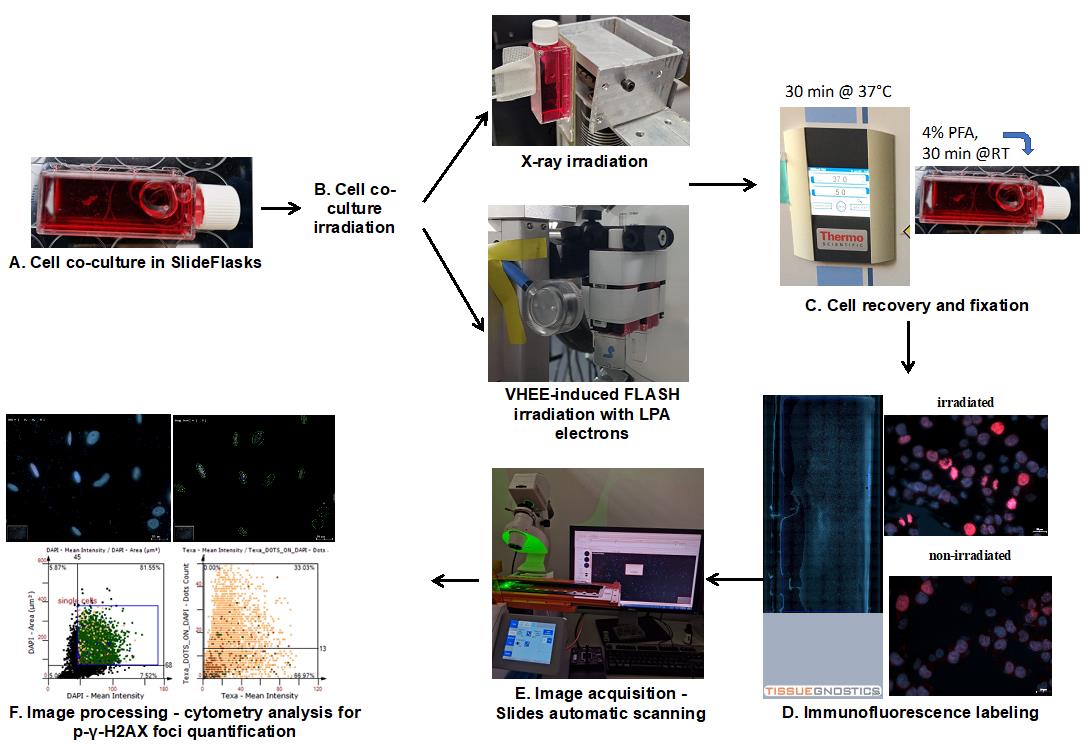
Workflow of cell co-culture irradiation and foci quantification analysis
Background
Very high energy electrons (VHEE), defined as electrons with energies between 150 and 250 MeV, were proposed as an alternative radiotherapy approach and found adequate to penetrate deep tumor sites [1]. High-intensity laser-driven acceleration of VHEE of dose rates as high as 1013 Gy/s is achievable by laser-plasma accelerators (LPA) with quasi-monoenergetic beams, being proposed as new platforms for VHEE-induced FLASH irradiation [2,3]. We have recently confirmed the cell DNA damage–producing capacity of the laser-driven high-energy dose electrons produced by a tabletop high-power LPA in cancer cells for future FLASH-radiotherapy applications [4].
In order to investigate the potential sparing effect of FLASH radiation on normal cells while evaluating the conditions where it induces enhanced DNA damage to cancer cells, we set up a protocol that allows simultaneous exposure of normal and malignant cells to irradiation by LPA-produced electrons for comparison. The golden standard method in the field of radiobiology to evaluate the effects of radiation on cells is by analyzing nuclear foci that accumulate at DNA damage sites [5,6]. The ionizing radiation is known to produce DNA double-strand breaks (DSBs) that affect preponderantly the cells engaged in the cell cycle [7]. Therefore, this type of radiation is used in radiotherapy to eliminate highly proliferative cancer cells from tumors [8]. Upon exposure of cells to radiation or other DNA-damaging agents, the DNA repair molecular mechanism is activated [7]. Proteins accumulating and forming foci at the sites of insult can be used as markers to quantitate the extent of the produced damage [6], as well as for monitoring the kinetics of DNA repair [9]. Ideally, the radiation regimen should have a maximal impact on cancer cells without affecting the normal function of other cells in the tissue. Using our proposed co-culture protocol, one can simultaneously observe the effects produced by a certain radiation dose and regimen on other cells in the target tissue alongside the cancer cells. This approach has several key advantages, namely 1) minimizing experiment-to-experiment variability when comparing the effect of a certain irradiation regime (type of source and dose) on different cell types; 2) allowing cell–cell communication between different cell types that modulate the outcome on each cell type, similarly to events produced within the tumor microenvironment; and 3) minimizing the time required for radiation exposure, thereby increasing feasibility of large experiments where several doses have to be applied in replicates in the same day on cells from the same batch. Our method allows discriminating between different cell types within the same sample based on morphological and fluorescence parameters (image cytometry). Here, we present a case where different cell types have different nuclear shapes [A375 cells have round nuclei, while normal human epidermal melanocytes (NHEMs) have elongated nuclei]. Nevertheless, TissueQuest enables users to separate cell populations based on protein markers’ expression or other features that can be quantified using fluorescent labeling.
For this, we have harnessed the power of quantitative imaging that allows precise evaluation of foci number with increased throughput on a statistically significant number of cells per sample. Using an automatic scanning system and image cytometry software, we have recently compared the effects of VHEE radiation and conventional X-ray in co-cultures of melanoma (A375 cells) and NHEMs [4]. Upon recovery and labeling of irradiated cells with antibodies against p-γ-H2AX and a nuclear stain (i.e., Hoechst), we have successfully shown that the accelerated VHEE can induce an increase in DNA damage foci count in cancer cells. Noteworthy, in a specific area of the co-culture monolayer, the effect is only observed in cancer cells, while spearing the NHEMs from stress [4].
The protocol presented here can be adopted by researchers studying the effect of any type of radiation on adherent cells, as well as by investigators who apply other types of DNA damage agents to cells in vitro (e.g., cisplatin). Also, the detailed presentation of the quantitative cytometry analysis strategy will be useful for TissueFAXS microscopy users to implement automatic scanning and analysis in their laboratories.
Materials and reagents
Biological materials
1. A375 human metastatic melanoma cells [American Type Culture Collection (ATCC), catalog number: CRL-1619)]. The cells should be grown to 70%–80% confluency in A375 culture media (see Recipe 1) and kept in a humidified environment at 37 °C, in the presence of 5% CO2, in a cell culture incubator
2. Normal human epidermal melanocytes (NHEMs) (Lonza, catalog number: CC-2504)
Reagents
1. High-glucose (HG) Dulbecco’s modified Eagle’s medium (DMEM) containing L-GlutaMAX (Gibco, catalog number: 61965-026)
2. Melanocyte growth basal medium 4 (MBM-4) (Lonza, catalog number: CC-3250)
3. Melanocyte growth medium 4 (MGM-4), Single Quots Supplements (Lonza, catalog number: CC-4435)
4. Fetal bovine serum (FBS) (Gibco, catalog number: 10437-028)
5. Penicillin-Streptomycin, 10,000 U/mL to 10,000 μg/mL (Gibco, catalog number: 15140122)
6. 1× Dulbecco's phosphate-buffered saline (DPBS) without calcium (Ca) and magnesium (Mg) (Gibco, catalog number: 14190250)
7. 0.05% Trypsin-EDTA solution (Gibco, catalog number: 25300054)
8. 0.025% Trypsin-EDTA solution (Lonza, catalog number: CC-5012)
9. 0.02% Versene (EDTA) solution (Lonza, catalog number: 17-711E)
10. Trypsin neutralizing solution (TNS) (Lonza, catalog number: CC-5002)
11. 0.4% Trypan Blue solution (StemCell Technologies, catalog number: 7050)
12. Bovine serum albumin (BSA) powder (Sigma-Aldrich, catalog number: A9647-10G)
13. Cisplatin powder (Santa Cruz, catalog number: sc-200896)
14. Mouse monoclonal antibodies for the Ser140 phosphorylated form of γ-H2AX (Invitrogen, Thermo, catalog number: MA1-2022)
15. Alexa Fluor 594-conjugated donkey anti-mouse secondary antibodies (Invitrogen, Thermo, catalog number: A21203)
16. Triton X-100 solution (Sigma-Aldrich, catalog number: T9284-500ML)
17. Paraformaldehyde (PFA) powder (Sigma-Aldrich, catalog number: P6148-500G)
18. Hoechst 33342 (Molecular Probes, Thermo, catalog number: H21492)
19. FluorSave Reagent (Millipore, catalog number: 345789)
20. NaOH (ADRA CHIM SRL, catalog number: 011-002-00-6)
Solutions
1. A375 cell culture media (complete DMEM HG) (see Recipes)
2. NHEM cell culture media (complete MBM-4) (see Recipes)
3. 10 mM cisplatin stock solution (see Recipes)
4. 5 M NaOH solution (see Recipes)
5. 4% PFA solution (see Recipes)
6. 0.2% Triton X-100 permeabilization solution (see Recipes)
7. 0.5% BSA blocking solution (see Recipes)
8. Primary antibody incubation solution (see Recipes)
9. Secondary antibody incubation solution (see Recipes)
10. Hoechst staining solution (see Recipes)
Recipes
1. A375 cell culture media
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DMEM HG medium | n/a | 445 mL |
| Heat-inactivated FBS | 10% | 50 mL (see note*) |
| Penicillin-streptomycin solution | 100 units/mL penicillin, 100 μg/mL streptomycin | 5 mL |
| Total (optional) | n/a | 500 mL |
*Note: Before preparing the media, inactivate the bottle of FBS into a water bath for 30 min at 56 °C. Allow the heat-inactivated FBS (hiFBS) solution to cool down before adding it to the complete DMEM HG media. Prepare hiFBS aliquots and store them in the -25 °C freezer.
2. NHEM cell culture media
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MBM-4 basal medium | n/a | 500 mL |
| MGM-4 Single Quots Supplements | n/a | 9 mL |
| Total (optional) | n/a | 509 mL |
*Note: After adding MGM-4 Single Quots Supplements to MBM-4 basal medium, use within one month. Do not re-freeze.
3. 10 mM cisplatin stock solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Cisplatin powder | n/a | 30 mg |
| MilliQ ultrapure water | n/a | Adjust to 10 mL |
| Total (optional) | 10 mM | 10 mL |
4. 5 M NaOH solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaOH pellets | n/a | 20 g |
| MilliQ ultrapure water | n/a | Adjust to 100 mL |
| Total (optional) | 5 M | 100 mL |
5. 4% PFA solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PFA powder | 4% | 2 g |
| PBS (see note *) | 1× | Adjust to 50 mL |
| NaOH solution | n/a | 3 μL sequential pipetting/dropwise until a clear solution is formed (~pH 7) |
| Total (optional) | n/a | 50 mL |
*Note: Heat 40 mL of PBS in a glass beaker under a fume hood to ~60 °C. Add the PFA powder and stir until it dissolves. Cool the solution to room temperature (RT) and then adjust the pH to 7 using NaOH. Bring the volume to 50 mL. Aliquot and store in the freezer.
6. 0.2% Triton X-100 permeabilization solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Triton X-100 solution (see note *) | as produced (0.2–0.9 mM) | 100 μL |
| PBS solution | 1× | 50 mL |
| Total (optional) | 0.2% | 50 mL |
*Note: Triton X-100 is a viscous solution. Cut the pipette tip before aspirating the solution. Release the tip in the PBS solution after resuspending it three times.
7. 0.5% BSA blocking solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| BSA powder | n/a | 0.25 g |
| PBS solution (see note *) | 1× | Adjust to 50 mL |
| Total (optional) | 0.5% | 50 mL |
*Note: Add small amounts of PBS solution onto the BSA powder and mix gently. Otherwise, the mixed solution will become foamy. If so, centrifuge at the maximum speed rotation allowed for the centrifuge tube in which you dissolved the powder.
8. Primary antibody incubation solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Mouse anti-p-γ-H2AX | 1 mg/mL | |
| BSA blocking solution | 0.5% | |
| Total (optional) | 1:200 |
9. Secondary antibody incubation solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| AlexaFluor 594-conjugated donkey anti-mouse secondary antibody | 2 mg/mL | |
| BSA blocking solution | 0.5% | |
| Total (optional) | 1:400 |
10. Hoechst staining solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Hoechst 33342 stock solution | 10 mg/mL | 1 μL |
| PBS solution | 1× | 3 mL |
| Total (optional) | 1:3,000 | 3 mL |
Laboratory supplies
1. T-75 cell culture flasks (Corning Life Sciences, catalog number: 430641U)
2. T-25 cell culture flasks (Corning Life Sciences, catalog number: 430639)
3. SlideFlasks (Thermo Scientific, catalog number: 734-2107); the package contains a chamber detachment tool
4. Sterile Corning 24-well polystyrene tissue culture plates (Corning Life Sciences, catalog number: 3526)
5. Sterile 15 mL polypropylene centrifuge tubes (Corning Life Sciences, catalog number: 430790)
6. Sterile 50 mL polypropylene centrifuge tubes (Corning Life Sciences, catalog number: 430828)
7. 2 mL serological pipettes (Corning Life Sciences, catalog number: 4486)
8. 5 mL serological pipettes (Corning Life Sciences, catalog number: 4487)
9. 10 mL serological pipettes (Corning Life Sciences, catalog number: 4488)
10. Microcentrifuge tubes (ratiolab, catalog number: 5616027)
11. 12 mm diameter coverslips (CS), circular (Marienfeld, catalog number: 0111520)
12. Microscope slides (Marienfeld, catalog number: 1000200)
13. Forceps (Millipore, catalog number: XX6200006P)
14. Syringe needles
15. Pipette tips (compatible with your automatic pipettes)
16. Microscope cover glasses for chamber slides (Nunc/Thermo, catalog number: 171080)
17. Marienfeld counting chambers (Neubauer, catalog number: MARI0640130)
18. Microscope cover glasses 22 × 22 mm for counting chamber (Paul Marienfeld GmbH & Co, Marienfeld SUPERIOR, catalog number: 0101050)
19. Carl Zeiss Immersol immersion oil for microscopy (Zeiss, catalog number: 518F)
Equipment
1. Cell culture incubator maintained at 37 °C and 5% CO2 (Thermo Scientific, model: Heracell VIOS 250i, catalog number: 13-998-253)
2. Laminar-flow biosafety cabinet (Thermo Scientific, model: KS 15, Class II)
3. Refrigerated centrifuge (Beckman Coulter, model: Allegra X-12R)
4. MilliQ water purification system (Millipore, Model SAS 67120 water purification with filter Biopak Polisher, catalog number: CDUFBI0A1)
5. Stripettor Ultra Pipet Controller (Corning, catalog number: 15536304)
6. Eppendorf Research plus P10 mechanical pipette (Eppendorf, catalog number: 3123000020)
7. Eppendorf Research plus P20 mechanical pipette (Eppendorf, catalog number: 3123000039)
8. Eppendorf Research plus P200 mechanical pipette (Eppendorf, catalog number: 3124000083
9. Eppendorf Research plus P1000 mechanical pipette (Eppendorf, catalog number: 3123000063)
10. X-ray system consisting of a NIKON XT H 225kV reflection target source with 3 μm focal spot size (Nikon Metrology NV) and a PTW UNIDOS T 10005-50406 Electromer connected to a Farmer ionization chamber TN30010 (calibrated at PTB Germany)
11. X-ray detector system: Gafchromic EBT3-810 dosimetry film (Ashland Global Holdings Inc., product code: 828204)
12. Customized chopper manufactured by selective laser melting (SLM) using 3D printing from steel with brass infusion. X-ray chopper specifications: 12 slits of 8 mm (height) × 1.5 mm (width) at 15° apart from each other
13. LPA system (Thales, France) based on a Ti:Sapphire high-intensity laser (800 nm, 25 fs pulse duration, 25 J energy at 0.1 Hz, 1 J at 10 Hz), beam transport (Ardop, France), interaction chamber (Astra, Gemini, RAL UK)
14. Electron beam detector systems: ionization chamber (model Advance Markus, type TN34045), three thermoluminescent dosimeters (model Panasonic), Gafchromic film (model GF-EBT3)
15. Automated imaging system (TissueGnostics, Vienna, Austria, model: TissueFAXSiPlus i12), based on the Zeiss Axio Observer.Z1 motorized inverted microscope appended with an ultra-precise motorized stage for automated sample acquisition with:
a. Oil immersion objective lens (Zeiss, model: Plan-Apochromat 63×/1.4 Oil DIC M27, 0.19 mm working distance)
b. Air objective lens (Zeiss, model: LD Plan-Neofluar 10×/0.3 M27, 5.3 mm working distance)
c. Air objective lens (Zeiss, model: LD Plan-Neofluar 20×/0.4 Korr M27, 7.9 mm working distance)
d. DAPI filter (excitation λ: 360 nm; emission λ: 462 nm), associated to the DAPI channel
e. Alexa 568/Cy3 filter (excitation λ: 568 nm; emission λ: 603 nm), associated to the TxRed channel
f. High-sensitivity digital monochrome camera for fluorescence microscopy (PCO AG, Kelheim, Germany; model: PCO PixelFly 14-bit dynamic range grayscale CCD camera; catalog number: C11440-10C)
g. Microscope fluorescence light source: Excelitas X-Cite 120PC Q (Cambridge Scientific, catalog number: 11419)
Note: Full instrument configuration is available at https://www.biochim.ro/facility-11/.
Note: Due to the necessity of recovering the cells in a CO2 incubator before fixation (and also keeping them in a physiological milieu immediately prior to the electrons exposure), the irradiation facility should be close to a tissue culture lab. Alternatively, for short periods of time (up to 4 h), cells could be kept at 37 °C in HEPES buffered media. This should be tested before the experiment because not all cell lines cope well in these conditions.
Software and datasets
1. TissueFAXS Slides Module (version 3.5.5.0129, release date 20 August 2012)
2. TissueQuest (version 4.0.1.0140, release date 18 November 2014)
3. Microsoft Office 365 Excel
4. GraphPad Prism (version 9.5.1 (528), release date 24 January 2023)
Procedure
文章信息
稿件历史记录
提交日期: Oct 14, 2024
接收日期: Dec 23, 2024
在线发布日期: Jan 19, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Orobeti, S., Dinca, I., Bran, A., Tiseanu, I., Sima, F., Petrescu, S. M. and Sima, L. E. (2025). Streamlined Quantification of p-γ-H2AX Foci for DNA Damage Analysis in Melanoma and Melanocyte Co-cultures Exposed to FLASH Irradiation Using Automated Image Cytometry. Bio-protocol 15(4): e5208. DOI: 10.21769/BioProtoc.5208.
- Orobeti, S., Sima, L. E., Porosnicu, I., Diplasu, C., Giubega, G., Cojocaru, G., Ungureanu, R., Dobrea, C., Serbanescu, M., Mihalcea, A., et al. (2024). First in vitro cell co-culture experiments using laser-induced high-energy electron FLASH irradiation for the development of anti-cancer therapeutic strategies. Sci Rep. 14(1): 14866.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
细胞生物学 > 基于细胞的分析方法 >
细胞生物学 > 基于细胞的分析方法 > 流式细胞术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link