- EN - English
- CN - 中文
Profiling the Secretome of Glioblastoma Cells Under Histone Deacetylase Inhibition Using Mass Spectrometry
利用质谱分析组蛋白去乙酰化酶抑制下胶质母细胞瘤细胞的分泌组特征
发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5197 浏览次数: 2053
评审: Nona FarbehiRupam GhoshAnonymous reviewer(s)

相关实验方案

适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
Basil Baby Mattamana [...] Peter Allen Faull
2025年12月20日 1261 阅读
Abstract
Glioblastoma (GBM) is the most aggressive brain tumor, and different efforts have been employed in the search for new drugs and therapeutic protocols for GBM. A label-free, mass spectrometry–based quantitative proteomics has been developed to identify and characterize proteins that are differentially expressed in GBM to gain a better understanding of the interactions and functions that lead to the pathological state focusing on the extracellular matrix (ECM). The main challenge in GBM research has been to identify novel molecular therapeutic targets and accurate diagnostic/prognostic biomarkers. To better investigate the GBM secretome upon in vitro treatment with histone deacetylase inhibitor (iHDAC), we employed a high-throughput label-free methodology of protein identification and quantification based on mass spectrometry followed by in silico studies. Our analysis revealed significant changes in the ECM protein profile, particularly those associated with the angiogenic matrisome. Proteins such as decorin, ADAM10, ADAM12, and ADAM15 were differentially regulated upon in silico analysis. In contrast, key angiogenesis markers such as VEGF and ECM proteins like fibronectin and integrins did not display significant changes. These results suggest that iHDAC inhibitors may modulate or suppress tumor behavior growth by targeting ECM proteins’ secretion rather than directly inhibiting angiogenesis.
Key features
• Analysis of the secretome of U87MG glioblastoma cells.
• Studies of mass spectrometry designed to modulate GBM biology and behavior focused on histone deacetylase inhibitors (iHDAC).
• Mass spectrometry was developed to identify and characterize proteins that are differentially expressed in GBM.
Keywords: Epigenetics (表观遗传学)Graphical overview
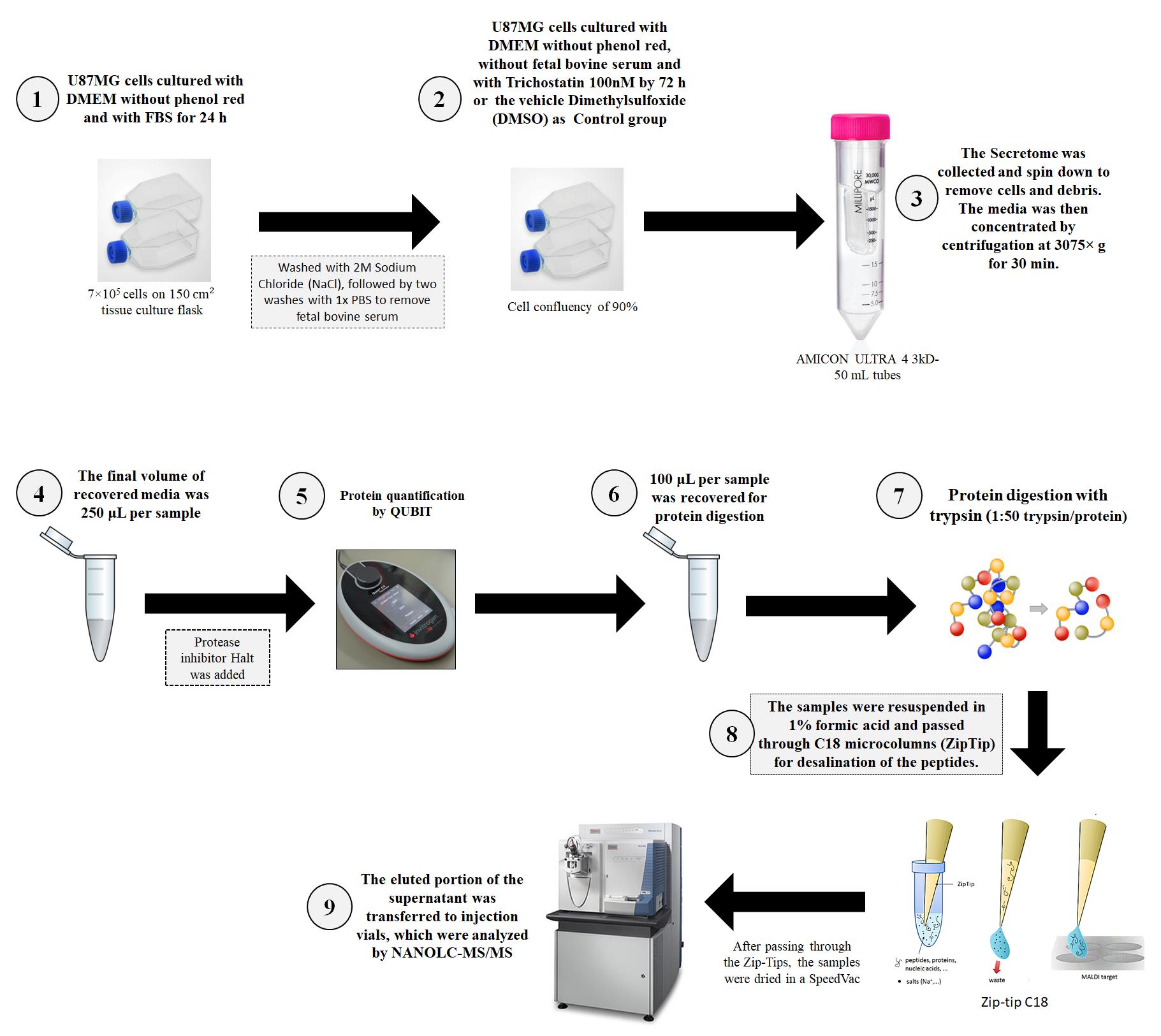
Background
The identification of novel molecular therapeutic targets and accurate diagnostic/prognostic biomarkers has been a major challenge in glioblastoma (GBM) research. Proteins have the potential to be used as diagnostic and prognostic biomarkers in patients with brain tumors. They can be detected in glioblastoma cells and in liquid matrices such as blood and its derivatives, cerebrospinal fluid (CSF), and urine [1]. Currently, the main approach in the search for tumor markers is the study of proteomic profiling, along with the study of gene expression [2]. In the repertoire of tumor-associated proteins, a great variety of functional proteins, peptides, and other biomolecules are secreted or released by tumor cells to promote abnormal cell growth, invasion, and metastasis. Some of these proteins include growth factors, chemokines, and angiogenic factors, resulting in ECM remodeling, modulation of cellular signaling, and inflammatory responses in the tumor microenvironment. These proteins belong to pathways classically related to alterations in the tumor microenvironment and, for this reason, might be eligible as biomarkers for tracking tumor growth or relapse in non-invasive strategies upon surgery, chemo, and radiotherapy [3,4]. In this sense, the tumor secretome may reflect tumor behavior and predict patient management. In this work, we studied the alterations in the GBM secretome upon the treatment of U87-MG cells with a histone deacetylase inhibitor (iHDAC). In fact, preclinical studies have demonstrated the efficiency of different iHDACs as antitumor agents, especially when associated with other therapies, including chemotherapy and radiotherapy [5]. The main mechanisms involved in iHDACs efficiency as anti-tumor drugs are related to the reduction in the expression of genes involved in DNA repair, mitotic spindle formation, homologous chromosome segregation, and positive modulation of apoptosis [6].
The translation of iHDAC inhibitors into clinical practice requires a deeper understanding of their mechanism of action. To advance the clinical application of iHDAC inhibitors, it is essential to understand their impact on the tumor mechanisms that shape their microenvironment. By combining epigenetic and biochemical profiling, we can uncover the link between iHDACs and secreted molecular factors that drive tumor growth and potentially lead to new therapeutic strategies [7].
To understand how iHDAC inhibitors influence glioblastoma (GBM) secretome, we examined the total set of proteins secreted by iHDAC-treated GBM cells using mass spectrometry. Our analysis revealed significant changes in the extracellular matrix (ECM) protein profile, particularly those associated with the angiogenic matrisome. Proteins such as decorin, ADAM10, ADAM12, and ADAM15 were differentially regulated upon in silico analysis. In contrast, key angiogenesis markers such as VEGF and the ECM proteins fibronectin and integrins did not display significant changes. These results indicate that iHDAC inhibitors may modulate tumor behavior by targeting the ECM proteins’ secretion, rather than directly inhibiting angiogenesis [7].
Here, we describe a step-by-step protocol for our GBM cell line secretome analysis protocol, further categorization of tumor-secreted proteins, and the characterization of pathways in which these proteins play already known roles.
Materials and reagents
Biological materials
1. U87MG glioblastoma cell line (Banco de Células do Rio de Janeiro, BCRJ Code: 0241)
Reagents
1. Dulbecco's modified Eagle's medium (DMEM) low glucose (Sigma-Aldrich, catalog number: D5523)
2. Fetal bovine serum (FBS) (LGC Biotecnologia, catalog number: 10BioPlus-500)
3. Penicillin and streptomycin (PS) (Sigma-Aldrich, catalog number: P4333)
4. DMEM low-glucose without red phenol (Sigma-Aldrich, catalog number: D2902)
5. Trichostatin A (TSA) (Sigma-Aldrich, catalog number: T1952)
6. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014)
7. Magnesium chloride (MgCl2·2H2O) (Sigma-Aldrich, catalog number: M8266)
8. Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: 223506)
9. Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P3911)
10. Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S9763)
11. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650)
12. HaltTM protease inhibitor cocktail 100× (Thermo Fisher, catalog number: 78429)
13. QubitTM Protein and Protein Broad Range (BR) Assay kits (Invitrogen, catalog number: Q33211)
14. Urea (Sigma-Aldrich, catalog number: U4884)
15. Thiourea (Sigma-Aldrich, catalog number: T8656)
16. HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] (Sigma-Aldrich, catalog number: 5310-OP)
17. DL-Dithiothreitol (DTT) powder (Sigma-Aldrich, catalog number: 43819-5g)
18. Iodoacetamide (IAA) powder (Sigma, catalog number: I1149-25g)
19. Sequencing-grade modified trypsin (Promega, catalog number: V5111), see the product information
20. Formic acid LC–MS grade (Thermo Fisher Scientific, catalog number: 28905)
21. Acetonitrile (ACN) (Sigma-Aldrich, catalog number: 900667)
Solutions
1. Phosphate buffer saline (PBS) (see Recipes)
2. NaCl 2 M solution (see Recipes)
3. Urea lysis buffer (see Recipes)
4. DTT solution (see Recipes)
5. IAA solution (see Recipes)
6. Solvent A (ACN 5% and formic acid 0.1%) (see Recipes)
7. Solvent B (ACN 95% and formic acid 0.1%) (see Recipes)
Recipes
1. Phosphate buffer saline 10× (PBS)
Note: Dissolve all reagents in 800 mL of H2O. Adjust the pH to 7.4 (or 7.2, if required) with HCl, and then add H2O to 1,000 mL.
| Reagent | Final concentration (10×) | Quantity or Volume |
|---|---|---|
| NaCl | 137 mM | 80 g |
| KCl | 2.7 mM | 2 g |
| Na2HPO4 | 10 mM | 14.4 g |
| KH2PO4 | 1.8 mM | 2.4 g |
| CaCl2·2H2O | 1 mM | 1.33 g |
| MgCl·2H2O | 0.5 mM | 1.0 g |
| H2O MilliQ | n/a | 800 mL |
| Total | n/a | 1,000 mL |
2. NaCl 2 M solution
Note: Dissolve 116.9 g of NaCl in approximately 800 mL of H2O MilliQ, then add more water until a final volume of 1,000 mL.
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 2 mol/L | 116.9 g |
| H2O MilliQ | n/a | 800 mL |
| Total | n/a | 1,000 mL |
3. Urea lysis buffer
Note 1: Dissolving urea is an endothermic reaction. The preparation can be facilitated by placing a stir bar in the beaker and using a warm water bath on a stir plate. Attention, this water should not be too hot! To prevent urea from precipitating, the buffer should be used at room temperature.
Note 2: The urea lysis buffer should be prepared before each experiment. Aliquots can be stored in the -80 °C freezer for up to 6 months.
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 7 mol/L | 42.04 g |
| Thiourea | 2 mol/L | 15.22 g |
| HEPES | 0.2 mol/L | 4.76 g |
| H2O MilliQ | n/a | 80 mL |
| Total | n/a | 100 mL |
4. DTT solution
Note: Dissolve 5 g of DTT in 324.14 mL of distilled water. After weighing the powder, store at -20 °C for up to one year. Thaw one aliquot for each experiment.
| Reagent | Final concentration | Amount |
|---|---|---|
| DTT | 100 mM | 5 g |
| H2O MilliQ | n/a | 324.14 mL |
5. Iodoacetamide (IAA) solution
Note: Dissolve 25 g of iodoacetamide in 338 mL of distilled water. After weighing the powder, store in the dark and add water only immediately before use. The iodoacetamide solution should be prepared just before use in each experiment.
| Reagent | Final concentration | Amount |
|---|---|---|
| Iodoacetamide | 400 mM | 25 g |
| H2O MilliQ | n/a | 338 mL |
6. Solvent A
| Reagent | Final concentration | Amount |
|---|---|---|
| Acetonitrile | 5% | 0.5 mL |
| Formic acid | 0.1% | 0.01 mL |
| H2O MilliQ | n/a | 10 mL |
7. Solvent B
| Reagent | Final concentration | Amount |
|---|---|---|
| Acetonitrile | 95% | 9.5 mL |
| Formic acid | 0.1% | 0.01 mL |
| H2O MilliQ | n/a | 10 mL |
Laboratory supplies
1. Pipettes (Eppendorf)
2. Tips (Eppendorf)
3. 50 mL Falcon centrifuge tubes, polypropylene, sterile (Corning, catalog number: 352070)
4. Microcentrifuge tube (Corning, Axygen, catalog number: MCT-150-C)
5. Becker
6. Water baths
7. Analytical Balance
8. 150 cm2 U-shaped canted neck cell culture flask with plug seal cap (Corning, catalog number: 430823)
9. Amicon Ultra-15 centrifugal filter unit (Meck Millipore, catalog number: UFC900324)
10. Pierce® C18 tips (87784 Pierce C18 Tips, 100 μL, 96 tips)
Equipment
1. Centrifuge
2. Centrifuge Heraeus Megafuge 8R (Thermo Fisher Scientific, catalog number: 75007214)
3. Sorvall centrifuge (Thermo Fisher Scientific, model: Lynx 4000)
4. CO2 incubator for cell culture (Thermo Fisher Scientific, model: 310 Series)
5. Speedvac (Thermo Fisher Scientific, model: SC210P1-15)
6. Thermomixer R with 1.5 mL block (Eppendorf, catalog number: Z605271)
7. -20 °C freezer (Thermo Fisher Scientific)
8. -80 °C freezer (Thermo Fisher Scientific)
9. Qubit protein assay kit, 500 assays (Thermo Fisher, catalog number: Q33238)
10. NanoLC (liquid chromatography) (Thermo Fisher Scientific, model: Easy1000)
11. Mass spectrometer (Thermo Fisher Scientific, model: Q ExactiveTM Plus)
12. Fused silica tubing (IDEX Health and Science, catalog number: FS-110)
13. PicoTipTM Emitter PicoFritTM SELF/P Capillary column (New Objective, catalog number: PF360-75-15-N-5)
14. Reprosil-Gold C18 (Dr. Maisch, catalog number: r33.9g-3g)
15. Reprosil-Pur C18 (Dr. Maisch, catalog number: r23.aq-2g)
Software and datasets
1. Software Xcalibur, Thermo Fisher Scientific, 2.2, 2011
2. Proteome Discoverer (PD), Thermo Fisher Scientific, 2.1, 2015
3. Uniprot database, 2017. Available in: https://www.uniprot.org/
4. All data have been deposited in ProteomeXchange Consortium via the PRIDE. Available at https://www.ebi.ac.uk/pride/ with the dataset identifier PXD022982 at PRIDE username: reviewer_pxd022982@ebi.ac.uk password: OBpIGRa2.
5. Microsoft Excel (Microsoft) or equivalent spreadsheet tool
Procedure
文章信息
稿件历史记录
提交日期: Sep 7, 2024
接收日期: Dec 17, 2024
在线发布日期: Jan 14, 2025
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Menezes, A., Martins, Y., Nogueira, F. C. S., De Abreu Pereira, D. and Carneiro, K. (2025). Profiling the Secretome of Glioblastoma Cells Under Histone Deacetylase Inhibition Using Mass Spectrometry. Bio-protocol 15(3): e5197. DOI: 10.21769/BioProtoc.5197.
分类
癌症生物学 > 血管生成 > 肿瘤微环境
系统生物学 > 蛋白质组学 > 分泌蛋白质组
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











