- EN - English
- CN - 中文
Simple and Fail-safe Method to Transform Miniprep Escherichia coli Strain K12 Plasmid DNA Into Viable Agrobacterium tumefaciens EHA105 Cells for Plant Genetic Transformation
将Miniprep制备的大肠杆菌K12菌株质粒DNA转化为可用于植物遗传转化的根癌农杆菌EHA105细胞的简单可靠方法
发布: 2025年01月05日第15卷第1期 DOI: 10.21769/BioProtoc.5174 浏览次数: 2375
评审: Mutinda SylviaAnonymous reviewer(s)
Abstract
Agrobacterium-mediated gene transformation method is a vital molecular biology technique employed to develop transgenic plants. Plants are genetically engineered to develop disease-free varieties, knock out unsettling traits for crop improvement, or incorporate an antigenic protein to make the plant a green factory for edible vaccines. The method’s robustness was validated through successful transformations, demonstrating its effectiveness as a standard approach for researchers working in plant biotechnology. It enables the introduction of foreign DNA into plant genomes. Conventionally, plant genetic transformation has relied on time-consuming, costly, and technically demanding procedures, such as electroporation and chimeric viruses or biolistic methods, which usually yield variable transformation efficiencies. This study presents a simple and fail-safe protocol that involves a modified freeze-thaw and heat-shock concoction method. This approach involves a streamlined plasmid miniprep procedure to isolate high-quality plasmid DNA from Escherichia coli K12 strain, followed by a target-specific transfer into A. tumefaciens EHA105 strain. The optimized method minimizes DNA degradation and maximizes uptake by Agrobacterium cells, making it a reproducible and accessible protocol for various genetic engineering applications. The transformation efficiency is consistently high, enhancing plasmid uptake while maintaining cell viability, requiring minimal specialized equipment and reagents. The proposed protocol offers significant advantages, including simplicity, reliability, and cost-effectiveness, positioning it as a valuable alternative to traditional techniques in the field of plant biotechnology.
Key features
• Uses liquid nitrogen as a proxy for freezing.
• Plasmid DNA from competent bacterial cells is extracted using a user-friendly high-copy isolation kit.
• A maximum of five consecutive days is sufficient to complete the procedures.
Keywords: Agrobacterium tumefaciens (根癌农杆菌)Graphical overview
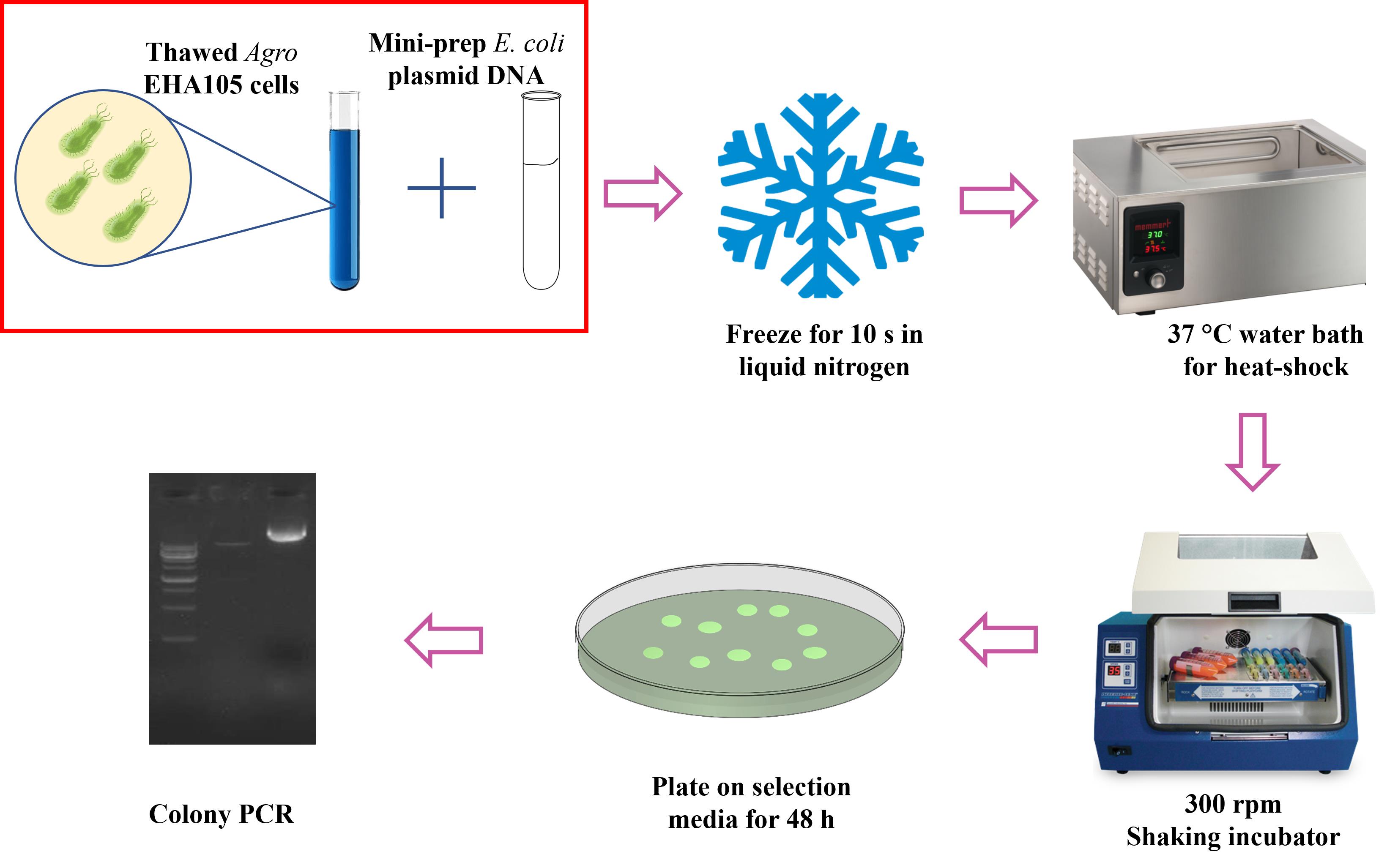
Freeze-thaw Agrobacterium transformation method
Background
Agrobacterium tumefaciens is a bacterium found in soil that has a natural ability for transient and stable transfer of foreign DNA into plant cells, including dicotyledonous and monocotyledonous species. This makes it a dominant technique for creating transgenic plants [1]. In the field of plant biotechnology, A. tumefaciens is widely employed to genetically modify plant genomes with desirable traits, such as enhanced nutritional content [2], disease resistance [3], and improved yield, or to vehicle plant-derived edible vaccines [4]. Traditional transformation methodologies using A. tumefaciens include electroporation, direct DNA transfer using chemically competent protocols, and the triparental mating method. Electroporation is a widely used approach, involving the application of an electrical pulse in the bacterial membrane to form pores for penetration of the DNA [5]. Despite being extremely efficient, it needs accurately optimized conditions and specialized equipment, limiting its accessibility for routine use [6]. Chemically competent methods, such as polyethylene glycol (PEG)-mediated transformation, are less sophisticated [7]; however, they present lower transformation efficiency in comparison to electroporation. The triparental mating technique, discovered by Herrera-Estrella et al. in 1983, involves the use of three strains, namely a donor, a helper, and a recipient [8]. It is highly effective yet time-consuming and very labor-intensive.
The freeze-thaw method simplifies the transformation process by eliminating the need for complex electroporation steps or triparental mating setup, making it more adaptable and accessible for various laboratory investigations [9]. The use of miniprep plasmid DNA directly from Escherichia coli reduces turnaround time, allowing for simple and rapid plasmid DNA isolation [10], coupled with uncompromised transformation efficiency [11]. This streamlined approach minimizes procedural errors and contamination chances that may occur when performing transformation experiments with more complex methodologies [12]. Additionally, the protocol is cost-effective, as it does not rely on expensive reagents or equipment; hence, it is easily reproducible and suitable, especially for laboratories with limited resources [13]. Also, it reports a low copy number of integrated transgenes, together with the possibility to transfer larger fragments of DNA. Nevertheless, the protocol is not without its limitations, as the transformation efficiency may be lower than that of optimized electroporation, particularly when dealing with complex gene constructs or larger plasmids [14]. Furthermore, the freeze-thaw method may need additional optimization when applied to different plasmid types or Agrobacterium strains, as variations in DNA uptake capabilities can influence transformation success [15]. Regardless of all these limitations, the protocol remains a practical alternative for routine transformation practices, especially in settings where simplicity and reliability are prioritized over maximal efficiency.
The protocol described in this article has broad utility and is a valuable tool in both basic and applied research across diverse areas of synthetic and microbial biotechnology [16]. It can be employed to introduce plasmids carrying reporter genes, selectable markers, and CRISPR/Cas9 components into A. tumefaciens, facilitating studies on gene expression, gene editing, and functional genomics in plants [17]. Similarly, the method can be extended to develop transgenic plants for biopharmaceutical production, environmental remediation, and biofortification.
Materials and reagents
The equipment, materials, and reagents listed below are appropriate for this protocol. Nonetheless, substitutes sourced elsewhere may be employed if they have shown comparable performance.
Biological materials
1. A. tumefaciens EHA105 strain (strains are preserved at -80 °C in our laboratory)
2. E. coli K12 strain (strains are preserved at -80 °C in our laboratory)
3. Plasmid DNA (stored at -80 °C in our laboratory)
Reagents
1. Lysogeny broth (LB) low salt (Sigma-Aldrich, catalog number: L3397-250G)
2. Agar powder, bacteriological grade (Himedia, catalog number: GRM026-500G)
3. Oligonucleotides (forward: AGGAAACAGCTATGACCATGATTACGAATTC, reverse: ACGTTGTAAAACGACGGCCAGTGCCAAGCTT)
4. OneTaq 2× Master Mix with standard buffer (New England Biolabs, catalog number: M0482)
5. 1 Kb Plus DNA marker (Invitrogen, catalog number: 10787-018)
6. SafeView gel stain (Applied Biological Materials, catalog number: G108)
7. DNA loading dye (Biolabs, catalog number: 10158560)
8. LE agarose (Cleaver Scientific, catalog number: 9012-36-6)
9. TAE buffer (Biological Industries, catalog number: 01-870-1A)
10. Nuclease-free water (BioConcept, catalog number: 3-07F04-H)
11. Rifampicin (GoldBio, catalog number: GB-R-120)
12. Kanamycin sulfate powder (Fisher Scientific, catalog number: BP 906-100)
13. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
Solutions
1. LB low-salt liquid medium (see Recipes)
2. LB low-salt agar solid medium (see Recipes)
3. 1% gel (see Recipes)
4. Rifampicin (see Recipes)
5. Kanamycin (see Recipes)
Recipes
1. LB low-salt liquid medium, pH 7.0
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Distilled water | n/a | 100 mL |
| LB low salt | 20 g/L | 2 g |
2. LB low-salt agar solid medium, pH 7.0
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Distilled water | n/a | 100 mL |
| LB low salt | 20 g/L | 2 g |
| Agar | 1.12% (w/v) | 1.12 g |
3. 1% gel
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| LE agarose | n/a | 0.5 g |
| TAE buffer | 1× | 50 mL |
| SafeView gel stain | n/a | 2 μL |
4. Rifampicin
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Rifampicin | 50 mg/mL | 0.5 g |
| DMSO | 99.9% | 10 mL |
5. Kanamycin
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Kanamycin sulfate | 50 mg/mL | 5 g |
| Distilled water | n/a | 100 mL |
Laboratory supplies
1. Petri plates (Borosil, catalog number: 3160072)
2. 500 mL culture bottle (Beckman, catalog number: 356011)
3. Powder-free nitrile gloves (Starlab, article number: SG-N-M)
4. Parafilm (Amor, catalog number: PM-996)
5. PCR reaction strips (Simport, catalog number: 330180784)
6. 10 μL pipette tips (AHN Biotechnologie, catalog number: P-214782)
7. 1,000 μL pipette tips (Biologix group, catalog number: MO002462ZA1230-12)
8. P10 micropipette (Eppendorf, catalog number: 4861000-0005)
9. P1000 micropipette (Eppendorf, catalog number: 4861000-0001)
10. 50 mL Falcon tubes (Biologix group, catalog number: 10-9502)
11. 1.5 mL microcentrifuge safe-lock tubes (Eppendorf, catalog number: 022363204)
12. Cell spreaders (ISOLAB, catalog number: CPSA20000002)
13. Isolate II Plasmid DNA kit (Bioline Meridian Biosciences, catalog number: BIO-52056)
14. Laboratory forceps (Thermo Fisher Scientific, catalog number: 112805)
15. Spectrophotometer cuvettes (Thermo Fisher Scientific, catalog number: 14-955-127)
16. Syringe microfilters 0.22 μm (GE Healthcare, catalog number: 289320100)
Equipment
1. Water bath (Cole-Parmer, model: WB-200)
2. Gel doc imaging (UVITEC Cambridge, model: UVIOOC H06)
3. Gel electrophoresis (Cleaver Scientific, model: NANOPAC300P)
4. Clean bench (Haier Biomedical, model: HCB-1600H)
5. Growth chamber (Daihan LabTech, model: LG C-5301)
6. Laboratory freezer (Haier Biomedical, catalog number: 0270501824)
7. Laboratory refrigerator (Haier Biomedical, catalog number: 0270501219A)
8. Thermal cycler (Eppendorf AG, model: 22331 Hamburg)
9. Biological safety cabinet (Haier Biomedical, model: HR1200-IIA2-S)
10. Shaking incubator (Winpact, model: SI-200)
11. Refrigerated microcentrifuge (Eppendorf, catalog number: 5430)
12. 50 mL high-efficient centrifuge (Hermile, catalog number: Z 446 K)
13. BioSpectrometer (Eppendorf, catalog number: 6135K1605057)
14. Laboratory water distiller (Liston, model: A 1210)
15. NanoDrop One (Thermo Fisher Scientific, model: 58595 Intertek)
16. Vertical high-pressure sterilization pot (Labnics, model: NVA-104)
17. Weighing balance (Mettler Toledo, model: PB1302-S/FACT)
18. pH meter (HANNA Instruments, model: H12211)
19. Variable speed vortex mixer (Thermo Fisher Scientific, catalog number: 9501200)
Software and datasets
1. Primer3 (4.1.0, 11/04/2017)
Procedure
文章信息
稿件历史记录
提交日期: Sep 13, 2024
接收日期: Dec 3, 2024
在线发布日期: Dec 18, 2024
出版日期: Jan 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Siamalube, B., Ehinmitan, E., Ngotho, M., Onguso, J. and Runo, S. (2025). Simple and Fail-safe Method to Transform Miniprep Escherichia coli Strain K12 Plasmid DNA Into Viable Agrobacterium tumefaciens EHA105 Cells for Plant Genetic Transformation. Bio-protocol 15(1): e5174. DOI: 10.21769/BioProtoc.5174.
分类
植物科学 > 植物转化 > 农杆菌介导的转化方法
生物科学 > 生物技术 > 微生物技术
植物科学 > 植物分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













