- EN - English
- CN - 中文
Protocol to Identify Unknown Flanking DNA Using Partially Overlapping Primer-based PCR for Genome Walking
基于部分重叠引物的PCR基因组步移技术用于未知侧翼DNA的鉴定
发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5172 浏览次数: 1379
评审: Sonali ChaturvediDavid PaulAnonymous reviewer(s)
Abstract
Genome walking is a popular molecular technique for accessing unknown flanking DNAs, which has been widely used in biology-related fields. Herein, a simple but accurate genome-walking protocol named partially overlapping primer (POP)-based PCR (POP-PCR) is described. This protocol exploits a POP set of three POPs to mediate genome walking. The three POPs have a 10 nt 3' overlap and 15 nt heterologous 5' regions. Therefore, a POP can partially anneal to the previous POP site only at a relatively low temperature (approximately 50 °C). In primary POP-PCR, the low-temperature (25 °C) cycle allows the primary POP to partially anneal to site(s) of an unknown flank and many sites of the genome, synthesizing many single-stranded DNAs. In the subsequent high-temperature (65 °C) cycle, the target single-stranded DNA is converted into double-stranded DNA by the sequence-specific primer, attributed to the presence of this primer complement, while non-target single-stranded DNA cannot become double-stranded because it lacks a binding site for both primers. As a result, only the target DNA is amplified in the remaining 65 °C cycles. In secondary or tertiary POP-PCR, the 50 °C cycle directs the POP to the previous POP site and synthesizes many single-stranded DNAs. However, as in the primary PCR, only the target DNA can be amplified in the subsequent 65 °C cycles. This POP-PCR protocol has many potential applications, such as screening microbes, identifying transgenic sites, or mining new genetic resources.
Key features
• This POP-PCR protocol, built upon the technique developed by Li et al. [1], is universal to genome walking of any species.
• The established protocol relies on the 10 nt 3' overlap among a set of three POPs.
• The first two rounds of POP-PCRs can generally give a positive walking outcome.
Keywords: Genome walking (基因组步移)Graphical overview
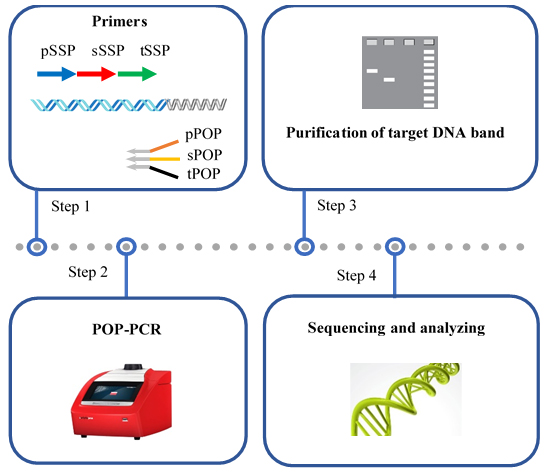
Background
In life science–related areas, genome walking is often employed to obtain unknown genomic DNA flanking known DNA [2–5]. Genome walking has significantly advanced areas such as genetics, biotechnology, and microbiology [6–9]. Many genome-walking methods have been proposed during the past four decades. Although the rationales or experimental steps involved are diverse, these methods can generally be classified into two groups: (i) genome treatment-dependent PCR and (ii) random PCR. The former requires digesting genomic DNA and then ligating the digested DNA with a cassette/adaptor/linker prior to PCR amplification. A random PCR, however, requires just a partial annealing of the walking primer to the unknown flank in the low-temperature (around 30 °C) cycle. A random PCR-based walking protocol has garnered much more attention than other protocols because it omits the time-consuming genome treatment process [10–14].
To date, approximately a dozen techniques of random PCR-based genome walking have been established, such as panhandle (or racket) PCR, thermal asymmetric interlaced PCR, and primer extension refractory PCR [6,12,15–18]. These techniques rely on the random annealing of the arbitrary walking primer to the unknown flanking genomic region, and the subsequent differential amplification between target DNA and non-target DNA. The reason for this differential amplification is that the annealing efficiency of the walking primer is lower than that of the paired sequence-specific primer (SSP). However, these techniques are still comprised of complex PCR steps or unsatisfactory amplification specificity [19–21]. Therefore, researchers have been actively attempting to devise a practical random PCR–based genome-walking technique.
Here, a novel genome-walking protocol of partially overlapping primer-based PCR (POP-PCR) is detailed for efficient genome walking. A POP-PCR set, comprising three rounds of nested amplifications, relies on the 10 nt 3' overlap among the three POPs to overcome non-target amplification. In each round of amplification, the POP has only one chance of partial annealing, as only one 25 °C or 50 °C cycle is performed in this amplification. Thus, only the target product can be amplified, while the non-target product is removed. In addition, unlike other walking methods, POP-PCR uses a unique POP for each round of amplification, which helps reduce non-target background arising from the other POPs. The proposed protocol can be used in many aspects of life science–related areas, like isolating promoters, unveiling new genetic resources, or identifying T-DNA [1,22–24].
Materials and reagents
Biological materials
1. Genomic DNA of rice, given by the Lab of Dr. Xiaojue Peng at Nanchang University
2. Genomic DNA of Pichia pastoris GS115, extracted using Dr. GenTLE (from Yeast) High Recovery kit (Takara, Dalian, China) according to the supplier’s instruction
3. Genomic DNA of Levilactobacillus brevis NCL912 [25–27], extracted using the method described by Li et al. [26]
4. Genomic DNA of human blood, extracted using the method described by Gustafson et al. [28]
Reagents
1. 10× LA PCR buffer (Mg2+ plus) (Takara, catalog number: RR042A)
2. 6× Loading buffer (Takara, catalog number: 9156)
3. LA Taq polymerase (hot-start version) (Takara, catalog number: RR042A)
4. dNTP mixture (Takara, catalog number: RR042A)
5. DL 5,000 DNA marker (Takara, catalog number: 3428Q)
6. 1× TE buffer (Sangon, catalog number: B548106)
7. Agarose (Sangon, catalog number: A620014)
8. 1 M NaOH (Yuanye, catalog number: B28412)
9. 0.5 M EDTA (Solarbio, catalog number: B540625)
10. Green fluorescent nucleic acid dye (10,000×) (Solarbio, catalog number: G8140)
11. Tris (Solarbio, catalog number: T8060)
12. Boric acid (Solarbio, catalog number: B8110)
13. Agarose Gel DNA Purification kit V2.0 (Takara, catalog number: DV805A)
14. Primers (Sangon)
pPOP1: AGTCAGCGTCCAGGTAGTCAGTCTC
sPOP1: TCAGGTCCAAGGTCAAGTCAGTCTC
tPOP1: CTCAGCGTGTTCGTCAGTCAGTCTC
pPOP2: CAGTCAGTCTCAGGTCGTCTCCAGT
sPOP2: AGCAGGTCAGTTACACGTCTCCAGT
tPOP2: TCAGTCAGTCAGTTGCGTCTCCAGT
pPOP3: CGCTTCAGATGGTACAGTGCAGTCA
sPOP3: ACACGATCCCAAGGTAGTGCAGTCA
tPOP3: GTTACTCAGGTCCCAAGTGCAGTCA
pPOP4: GCCTTGAACTGGACCTGATCGACTG
sPOP4: CATGACCGTGCTGAGTGATCGACTG
tPOP4: TGGACTGTGCTACCTTGATCGACTG
gadA-SSP1(5'): CATTTCCATAGGTTGCTCCAAGGTC
gadA-SSP2(5'): ACGTCATCTCAGTTGTTAGCCAACC
gadA-SSP3(5'): AGCCGGTTTGCTTTCAAATGATTCT
gadA-SSP1(3'): TGCGGATACTGATAACAAGACGACA
gadA-SSP2(3'): GGATTGAGAAAGAACGTACGGGTGA
gadA-SSP3(3'): TCCTGCATATCGGTAACGCCCAATC
ALDOA-SSP1(5'): AAATGCTGCAGCCTCCCTCTCACCC
ALDOA-SSP2(5'): AATACCAGAAATGTGCCCTCCCGTG
ALDOA-SSP3(5'): TGAGCTGGCAGGTTGTAGTCTCTGT
ALDOA-SSP1(3'): CCCTCGGACGATTGGACCTAGCTTG
ALDOA-SSP2(3'): GGTCTAACGGTGCCTCTCAGCCTCT
ALDOA-SSP3(3'): TCTGCCCTTCCCCATGGACGTAAGT
malQ-SSP1(5'): CTTCCTGGGTAAGCGTCAGCGTGTG
malQ-SSP2(5'): CAGCTTCGTCGGTAGATTGAACGCT
malQ-SSP3(5'): GGTGGTCAGCAGCCAGCTATATTCG
malQ-SSP1(3'): CGTCATCGCTGTATGGTGATTGGTG
malQ-SSP2(3'): CGGTGTTTACTCCTACAAAGTGCTC
malQ-SSP3(3'): GCTACATTGCCGACAGTAACAGTGC
hyg-SSP1(5'): CGGCAATTTCGATGATGCAGCTTGG
hyg-SSP2(5'): CGGGACTGTCGGGCGTACACAAATC
hyg-SSP3(5'): GACCGATGGCTGTGTAGAAGTACTC
hyg-SSP1(3'): AACTCCCCAATGTCAAGCACTTCCG
hyg-SSP2(3'): GAAACCATCGGCGCAGCTATTTACC
hyg-SSP3(3'): GAAAGCACGAGATTCTTCGCCCTCC
Solutions
1. 2.5× TBE buffer (see Recipes)
2. 0.5× TBE buffer (see Recipes)
3. 100 μM primer (see Recipes)
4. 10 μM primer (see Recipes)
5. 1.5% agarose gel (see Recipes)
Recipes
1. 2.5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 0.5 M EDTA solution | 5 mM | 10 mL |
| Tris | 225 mM | 27 g |
| Boric acid | 225 mM | 13.75 g |
| ddH2O | n/a | 950 mL |
| Total | n/a | 1,000 mL |
Adjust the pH to 8.3 with 1 M NaOH and then replenish the solution to 1,000 mL with ddH2O. This buffer can be stored at room temperature for three months.
2. 0.5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 2.5× TBE buffer | 0.5× | 200 mL |
| ddH2O | n/a | 800 mL |
| Total | n/a | 1,000 mL |
This buffer can be stored at room temperature for three months.
3. 100 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Powdery primer | 100 μM | n/a |
| 1× TE buffer | 1× | Volume specified in the sheet of primer synthesis |
| Total | n/a | Volume specified in the sheet of primer synthesis |
Note: Dilute a portion of the 100 μM primer to prepare 10 μM primer and store the remaining portion at -80 °C.
4. 10 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 100 μM primer | 10 μM | 1 μL |
| 1× TE buffer | 1× | 9 μL |
| Total | n/a | 10 μL |
Note: Prepare extra volume of a 10 μM primer and divide it into multiple 1.5 mL microcentrifuge tubes; then, store the microcentrifuge tubes at -80 °C. Take one tube at a time and store it at -20 °C after use.
5. 1.5% agarose gel
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1.5% | 1.5 g |
| 0.5× TBE buffer | 0.5× | 100 mL |
| Green fluorescent nucleic acid dye (10,000×) | 1× | 10 μL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 0.2 mL thin-wall PCR tubes (Kirgen, catalog number: KG2311)
2. 10 μL pipette tips (Sangon, catalog number: F600215)
3. 200 μL pipette tips (Sangon, catalog number: F600227)
4. 1,000 μL pipette tips (Sangon, catalog number: F630101)
5. 1.5 mL microcentrifuge tubes (Labselect, catalog number: MCT-001-150)
Equipment
1. PCR apparatus (Applied Biosystems, model: GeneAmp PCR System 2700)
2. Electrophoresis apparatus (Beijing Liuyi, model: DYY-6C)
3. Gel imaging system (Bio-Rad, model: ChemiDoc XRS+)
4. Microcentrifuge (Tiangen, model: TGear)
Software and datasets
1. Oligo 7 software (Molecular Biology Insights, Inc., USA)
2. DNASTAR Lasergene software (DNASTAR, Inc., USA)
Procedure
文章信息
稿件历史记录
提交日期: Sep 13, 2024
接收日期: Dec 2, 2024
在线发布日期: Dec 17, 2024
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Jia, M., Ding, D., Liu, X. and Li, H. (2025). Protocol to Identify Unknown Flanking DNA Using Partially Overlapping Primer-based PCR for Genome Walking. Bio-protocol 15(3): e5172. DOI: 10.21769/BioProtoc.5172.
分类
分子生物学 > DNA > PCR
微生物学 > 微生物遗传学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













