- EN - English
- CN - 中文
Protocol to Retrieve Unknown Flanking DNA Using Fork PCR for Genome Walking
利用叉式PCR进行基因组步移以扩增未知侧翼DNA的实验方案
发布: 2025年01月20日第15卷第2期 DOI: 10.21769/BioProtoc.5161 浏览次数: 2406
评审: Alba BlesaFernando A Gonzales-ZubiateAnonymous reviewer(s)
Abstract
PCR-based genome walking is one of the prevalent techniques implemented to acquire unknown flanking genomic DNAs. The worth of genome walking includes but is not limited to cloning full-length genes, mining new genes, and discovering regulatory regions of genes. Therefore, this technique has advanced molecular biology and related fields. However, the PCR amplification specificity of this technique needs to be further improved. Here, a practical protocol based on fork PCR is proposed for genome walking. This PCR uses a fork primer set of three arbitrary primers to execute walking amplification task, where the primary fork primer mediates walking by partially annealing to an unknown flank, and the fork-like structure formed between the three primers participates in inhibiting non-target amplification. In primary fork PCR, the low-annealing temperature (25 °C) cycle allows the primary fork primer to anneal to many sites of the genome, synthesizing a cluster of single-stranded DNAs; the subsequent 65 °C cycle processes the target single-strand into double-strand via the site-specific primer; then, the remaining 65 °C cycles selectively enrich this target DNA. However, any non-target single-stranded DNA formed in the 25 °C cycle cannot be further processed in the following 65 °C cycles because it lacks an exact binding site for any primer. Secondary, or even tertiary nested fork PCR further selectively enriches the target DNA. The practicability of fork PCR was validated by walking three genes in Levilactobacillus brevis CD0817 and one gene in Oryza sativa. The results indicated that the proposed protocol can serve as a supplement to the existing genome walking protocols.
Key features
• This protocol builds upon the method developed by Pan et al. [1], which is applicable to genome-walking for any species.
• The developed protocol is a random priming PCR-based genome-walking scheme.
• Two rounds of nested fork PCR amplifications suffice to release a positive walking result.
Keywords: Genome walking (基因组步移)Graphical overview
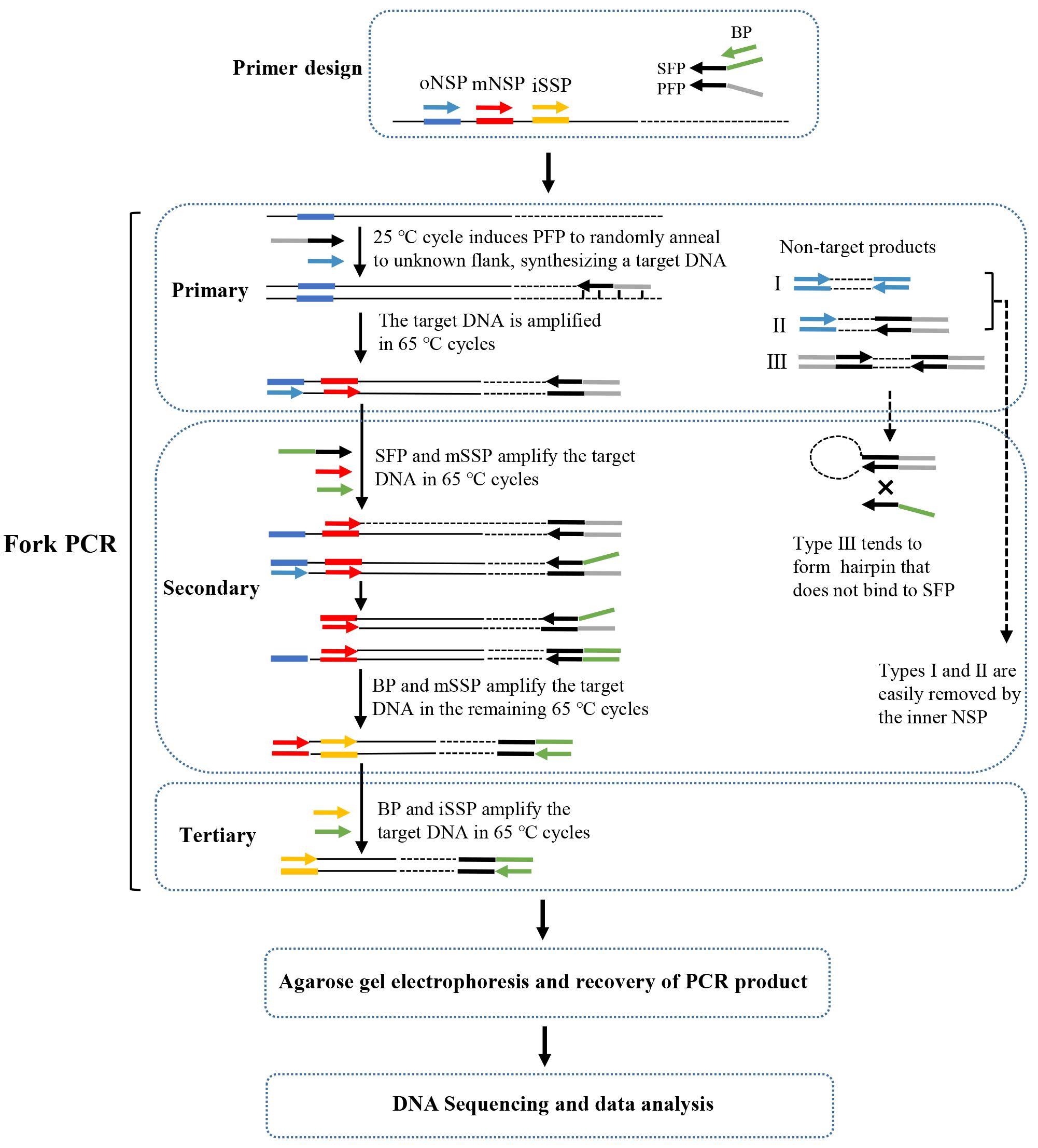
Background
Genome walking refers to a strategy used to mine unknown genomic regions flanking known DNAs. Genome walking has promoted the development of molecular biology–related fields by being widely applied, such as in cloning full-length genes, uncovering transgenic sites, and acquiring regulatory regions of genes. To date, many PCR-based genome walking techniques have been developed [2–6]. These techniques fall into two clusters according to the involved methodological principles: cluster I, genome pre-processing-dependent PCR [7–10], and cluster II, random priming PCR [11–13].
A random priming PCR method omits the genome pre-processing prior to PCR, having thus attracted increasing interest from researchers [14–16]. Over the past decades, about a dozen random PCR methods have been constructed, such as wristwatch PCR [4,17], racket PCR [3,5], and thermal asymmetric interlaced PCR [18,19]. In such a PCR, an arbitrary primer mediates genome walking by partially hybridizing to an unknown flank genomic region in the low-temperature cycle, while a nested sequence-specific primer(s) (NSP) ensures PCR amplification specificity. In general, this method requires at least two rounds of nested amplifications to obtain a target amplicon(s) but still suffers from a non-target background arising from the arbitrary primer [4,14]. Therefore, developing a random PCR genome walking method with satisfactory amplification specificity remains of interest.
Recently, we have developed a new genome-walking technique called fork PCR. The fork PCR depends on the partial overlaps between the three arbitrary primers [primary fork primer (PFP), secondary fork primer (SFP), and branch primer (BP)] in a fork primer set. PFP and SFP share a 3' part (stem), while their 5' parts (branches) are heterologous to each other. BP corresponds to the 5' branch of SFP. Therefore, PFP, SFP, and BP form a fork-like structure (Figure 1). The fork PCR has shown satisfactory amplification specificity mainly due to the following two facts: First, all thermal cycles in secondary/tertiary fork PCR are stringent (annealing temperature 65 °C). Second, the non-target product defined by PFP is hard to amplify in secondary/tertiary PCR as it tends to form a hairpin via intra-strand annealing mediated by the inverted PFP terminal repeats, rather than being annealed by SFP or BP. The feasibility of fork PCR has been verified by walking several selected genetic sites [1].
Materials and reagents
Biological materials
1. Genomic DNA of Levilactobacillus brevis CD0817 [20–23], prepared by our lab at Nanchang University (Nanchang, China)
2. Genomic DNA of Oryza sativa, obtained from the Lab of Dr. Xiaojue Peng at Nanchang University (Nanchang, China)
Reagents
1. LA Taq polymerase (hot-start version) (Takara, catalog number: RR042A)
2. dNTP mixture (Takara, catalog number: RR042A)
3. 10× LA PCR buffer (Mg2+ plus) (Takara, catalog number: RR042A)
4. 6× Loading buffer (Takara, catalog number: 9156)
5. DL 5,000 DNA marker (Takara, catalog number: 3428Q)
6. 1× TE buffer (Sangon, catalog number: B548106)
7. Agarose (Sangon, catalog number: A620014)
8. 1 M NaOH (Yuanye, catalog number: B28412)
9. Green fluorescent nucleic acid dye (10,000×) (Solarbio, catalog number: G8140)
10. 0.5 M EDTA (Solarbio, catalog number: B540625)
11. Boric acid (Solarbio, catalog number: B8110)
12. Tris (Solarbio, catalog number: T8060)
13. DiaSpin DNA Gel Extraction kit (Sangon, catalog number: B110092)
14. Primers (Sangon)
PFP1: 5'-ACGCGTAATAGCTCGGGATGATGCTGCTCGTGGATGACTCT-3'
SFP1: 5'-CCTGACCGCCTTCTACACCTATGCTGCTCGTGGATGACTCT-3'
PFP2: 5'-ATCCGCCCATAGCCTTCAGTGACTACGCTGCCTTGCTACTT-3'
SFP2: 5'-CCTGACCGCCTTCTACACCTGACTACGCTGCCTTGCTACTT-3'
BP: 5'-CCTGACCGCCTTCTACACCT-3'
oNSP-gadA: 5'-GTTTCTGGTCACAAGTACGGCATGG-3'
mNSP-gadA: 5'-TGCTGATACGCTGCCAGAAGAAATG-3'
iNSP-gadA: 5'-ACGGTTGACTCCATTGCCATTAACT-3'
oNSP-gadR: 5'-TCCTTCGTTCTTGATTCCATACCCT-3'
mNSP-gadR: 5'-CCATTTCCATAGGTTGCTCCAAGG-3'
iNSP-gadR: 5'-GGATACTGGCTAAAATGAATTAACTCGGATAA-3'
oNSP-pct: 5'-TCTTGTTCTTCAACAGTGGTGGGTA-3'
mNSP-pct: 5'-TCGTCTTTCGTGTAAGTGTTGGTGT-3'
iNSP-pct: 5'-AGGAAATATGCACTCTTGGGAAGCG-3'
oNSP-hyg: 5'-ACGGCAATTTCGATGATGCAGCTTG-3'
mNSP-hyg: 5'-GGGACTGTCGGGCGTACACAA-3'
iNSP-hyg: 5'-CTGGACCGATGGCTGTGTAGAAG-3'
Solutions
1. 2.5× TBE buffer (see Recipes)
2. 0.5× TBE buffer (see Recipes)
3. 100 μM primer (see Recipes)
4. 10 μM primer (see Recipes)
5. 1% agarose gel (see Recipes)
Recipes
1. 2.5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 0.5 M EDTA solution | 5 mM | 10 mL |
| Tris | 225 mM | 27 g |
| Boric acid | 225 mM | 13.75 g |
| ddH2O | n/a | 950 mL |
| Total | n/a | 1,000 mL |
Adjust pH to 8.3 with 1 M NaOH and then top the solution to 1,000 mL with ddH2O.
2. 0.5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 2.5× TBE buffer | 0.5× | 200 mL |
| ddH2O | n/a | 800 mL |
| Total | n/a | 1,000 mL |
3. 100 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Powdery primer | 100 μM | n/a |
| 1× TE buffer | 1× | Volume specified in the sheet of primer synthesis |
| Total | n/a | Volume specified in the sheet of primer synthesis |
Note: Dilute a portion of the 100 μM primer to prepare 10 μM primer and store the remaining portion at -80 °C.
4. 10 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 100μM primer | 10 μM | 1 μL |
| 1× TE buffer | 1× | 9 μL |
| Total | n/a | 10 μL |
Note: Prepare extra volume of a 10 μM primer and pipette it to multiple 1.5 mL microcentrifuge tubes. Then, store the tubes at -80 °C. Take one tube at a time and store it at -20 °C after use.
5. 1% agarose gel
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1% | 1 g |
| 0.5× TBE buffer | 0.5× | 100 mL |
| Green fluorescent nucleic acid dye (10,000×) | 1× | 10 μL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 0.2 mL thin-wall PCR tubes (Kirgen, catalog number: KG2311)
2. 10 μL pipette tips (Sangon, catalog number: F600215)
3. 200 μL pipette tips (Sangon, catalog number: F600227)
4. 1,000 μL pipette tips (Sangon, catalog number: F630101)
5. 1.5 mL microcentrifuge tubes (Labselect, catalog number: MCT-001-150)
Equipment
1. PCR apparatus (Analtytikjena, model: Biometra TAdvanced)
2. Microcentrifuge (Tiangen, model: TGear)
3. Electrophoresis apparatus (Beijing Liuyi, model: DYY-6C)
4. Gel imaging system (Bio-Rad, model: ChemiDoc XRS+)
Software and datasets
1. Oligo 7 software (Molecular Biology Insights, Inc., USA)
2. DNASTAR Lasergene software (DNASTAR, Inc.)
Procedure
文章信息
稿件历史记录
提交日期: Sep 14, 2024
接收日期: Nov 17, 2024
在线发布日期: Nov 27, 2024
出版日期: Jan 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Wu, H., Pan, H. and Li, H. (2025). Protocol to Retrieve Unknown Flanking DNA Using Fork PCR for Genome Walking. Bio-protocol 15(2): e5161. DOI: 10.21769/BioProtoc.5161.
分类
分子生物学 > DNA > PCR
微生物学 > 微生物遗传学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













